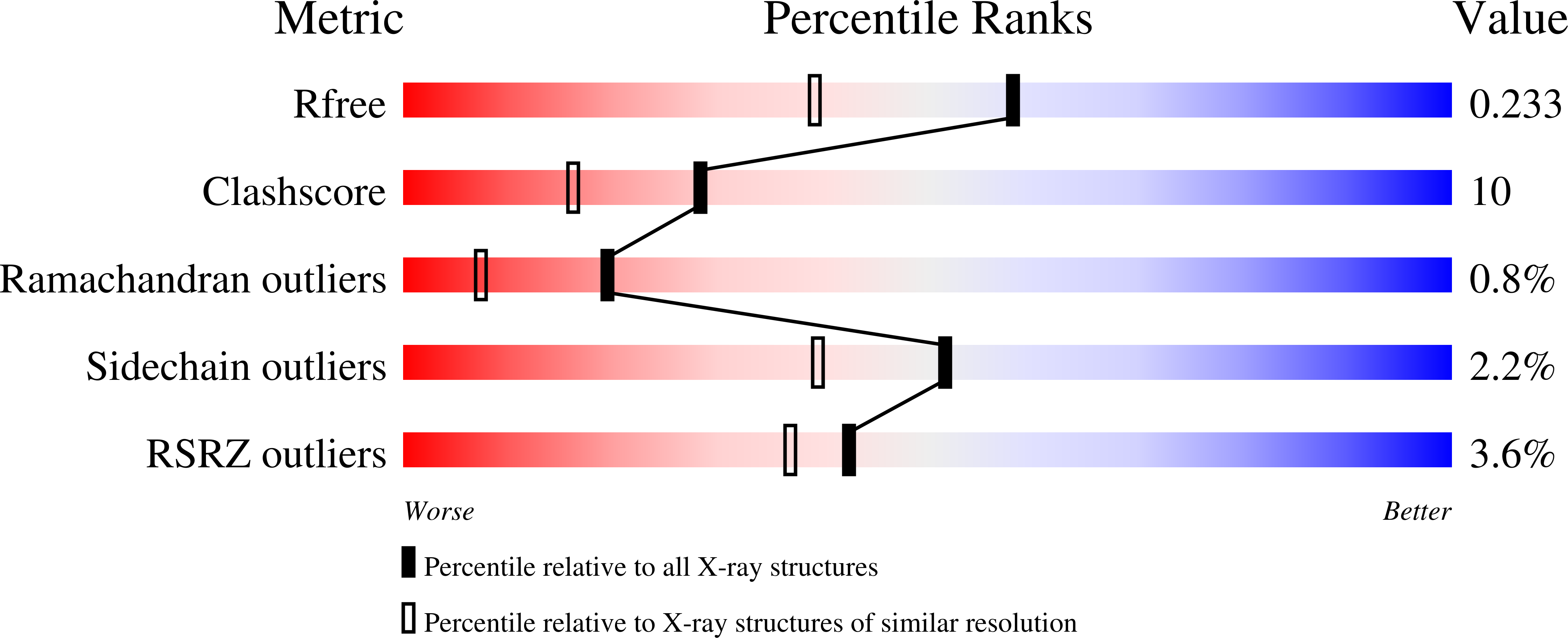
Crystal structure of hereditary vitamin D-resistant rickets--associated mutant H305Q of vitamin D nuclear receptor bound to its natural ligand
Rochel, N., Hourai, S., Moras, D.(2010) J Steroid Biochem Mol Biol 121: 84-87
- PubMed: 20403435
- DOI: https://doi.org/10.1016/j.jsbmb.2010.04.008
- Primary Citation of Related Structures:
3M7R - PubMed Abstract:
In the nuclear receptor of vitamin D (VDR) histidine 305 participates to the anchoring of the ligand. The VDR H305Q mutation was identified in a patient who exhibited the hereditary vitamin D-resistant rickets (HVDRR). We report the crystal structure of human VDR H305Q-ligand binding domain bound to 1alpha,25(OH)2D3 solved at 1.8A resolution. The protein adopts the active conformation of the wild-type liganded VDR. A local conformational flexibility at the mutation site weakens the hydrogen bond between the 25-OH with Gln305, thus explaining the lower affinity of the mutant proteins for calcitriol. The structure provides the basis for a rational approach to the design of more potent ligands for the treatment of HVDRR.
Organizational Affiliation:
IGBMC (Institut de G¨¦n¨¦tique et de Biologie Mol¨¦culaire et Cellulaire), D¨¦partement de Biologie et de G¨¦nomique Structurales, Centre National de la Recherche Scientifique, Institut National de la Sant¨¦ de la Recherche M¨¦dicale, Universit¨¦ de Strasbourg, 1 rue Laurent Fries, Illkirch, France. rochel@igbmc.fr