Crystal Structure Of I83E Meditope-Enabled Trastuzumab With Azido-Meditope
Williams, J.C., Bzymek, K.P., Pucket, J., Avery, K.A., Ma, Y., Xie, J., Zer, C., Horne, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MEMAB TRASTUZUMAB, LIGHT CHAIN | 214 | Homo sapiens | Mutation(s): 0 | 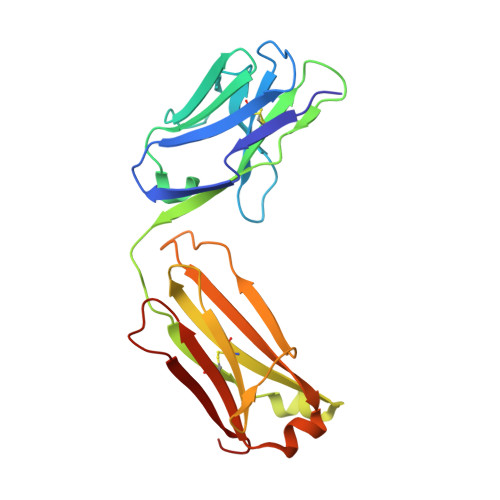 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MEMAB TRASTUZUMAB, HEAVY CHAIN | 223 | Homo sapiens | Mutation(s): 0 | 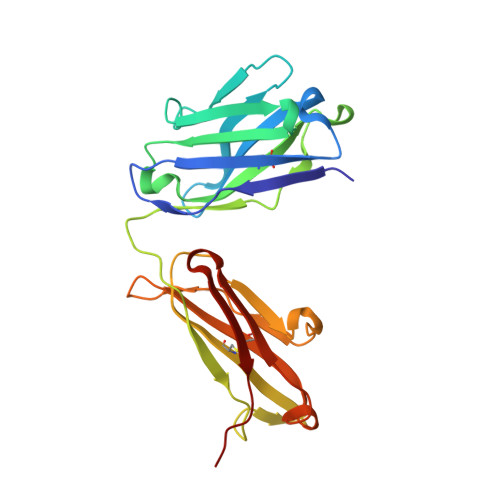 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein L | C [auth E] | 63 | Finegoldia magna | Mutation(s): 5 | 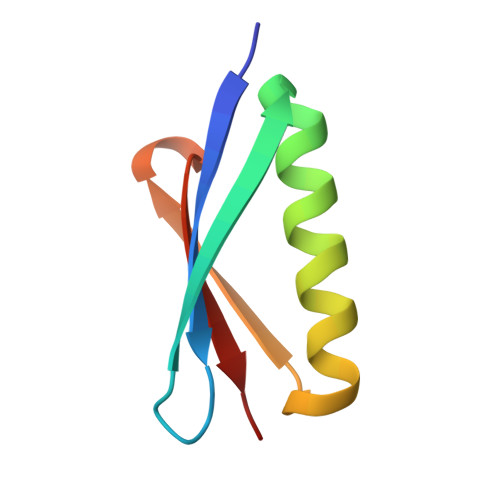 |
UniProt | |||||
Find proteins for Q51918 (Finegoldia magna) Explore Q51918 Go to UniProtKB: Q51918 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q51918 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Immunoglobulin G binding protein A | D [auth C] | 54 | Staphylococcus aureus | Mutation(s): 0 Gene Names: spa | 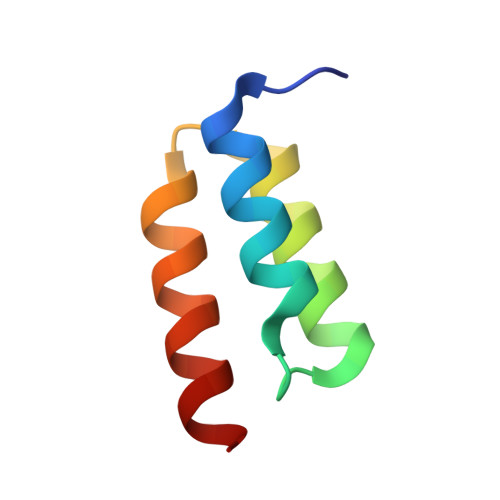 |
UniProt | |||||
Find proteins for P02976 (Staphylococcus aureus (strain NCTC 8325 / PS 47)) Explore P02976 Go to UniProtKB: P02976 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02976 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| AZIDO-PEG4-MEDITOPE | E [auth D] | 13 | synthetic construct | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MRY Query on MRY | F [auth B] | MESO-ERYTHRITOL C4 H10 O4 UNXHWFMMPAWVPI-ZXZARUISSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| 2GX Query on 2GX | E [auth D] | L-PEPTIDE LINKING | C15 H15 N O2 |  | PHE |
| Length ( ? ) | Angle ( ? ) |
|---|---|
| a = 53.53 | ¦Á = 90 |
| b = 105.47 | ¦Â = 90 |
| c = 117.13 | ¦Ă = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| MOLREP | phasing |
| PHENIX | refinement |