Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
Kremling, V., Sprenger, J., Oberthuer, D., Chapman, H.N., Middendorf, P., Falke, S., Kiene, A., Klopprogge, B., Scheer, T.E.S., Creon, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 2'-O-methyltransferase nsp16 | 304 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b EC: 2.1.1.57 | 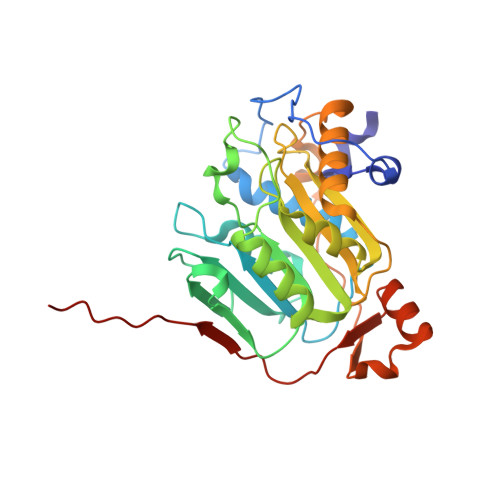 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Non-structural protein 10 | 140 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b | 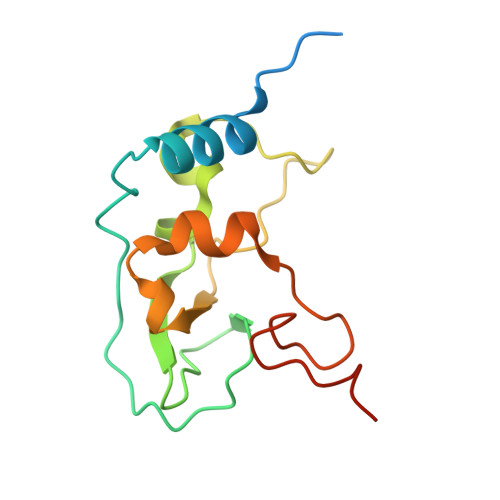 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1IO1 (Subject of Investigation/LOI) Query on A1IO1 | W [auth A] | (3~{S})-3-azanyl-4-[(3~{R},4~{R},6~{S})-3-[1,3-dimethyl-2,6-bis(oxidanylidene)purin-7-yl]-4-methyl-4,6-bis(oxidanyl)azepan-1-yl]-4-oxidanylidene-butanoic acid C18 H26 N6 O7 PXCRTYLWTBRTOT-JXKOVVAVSA-N |  | ||
| SAM Query on SAM | X [auth A] | S-ADENOSYLMETHIONINE C15 H22 N6 O5 S MEFKEPWMEQBLKI-FCKMPRQPSA-N |  | ||
| MES Query on MES | D [auth A] | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID C6 H13 N O4 S SXGZJKUKBWWHRA-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | BA [auth B], CA [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | AA [auth A] C [auth A] DA [auth B] E [auth A] EA [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CL Query on CL | FA [auth B], Y [auth A], Z [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( ? ) | Angle ( ? ) |
|---|---|
| a = 168.01 | α = 90 |
| b = 168.01 | β = 90 |
| c = 51.74 | γ = 120 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XDS | data scaling |
| PHENIX | refinement |
| Coot | model building |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | DESY Strategy Fund MUXCOSDYN |