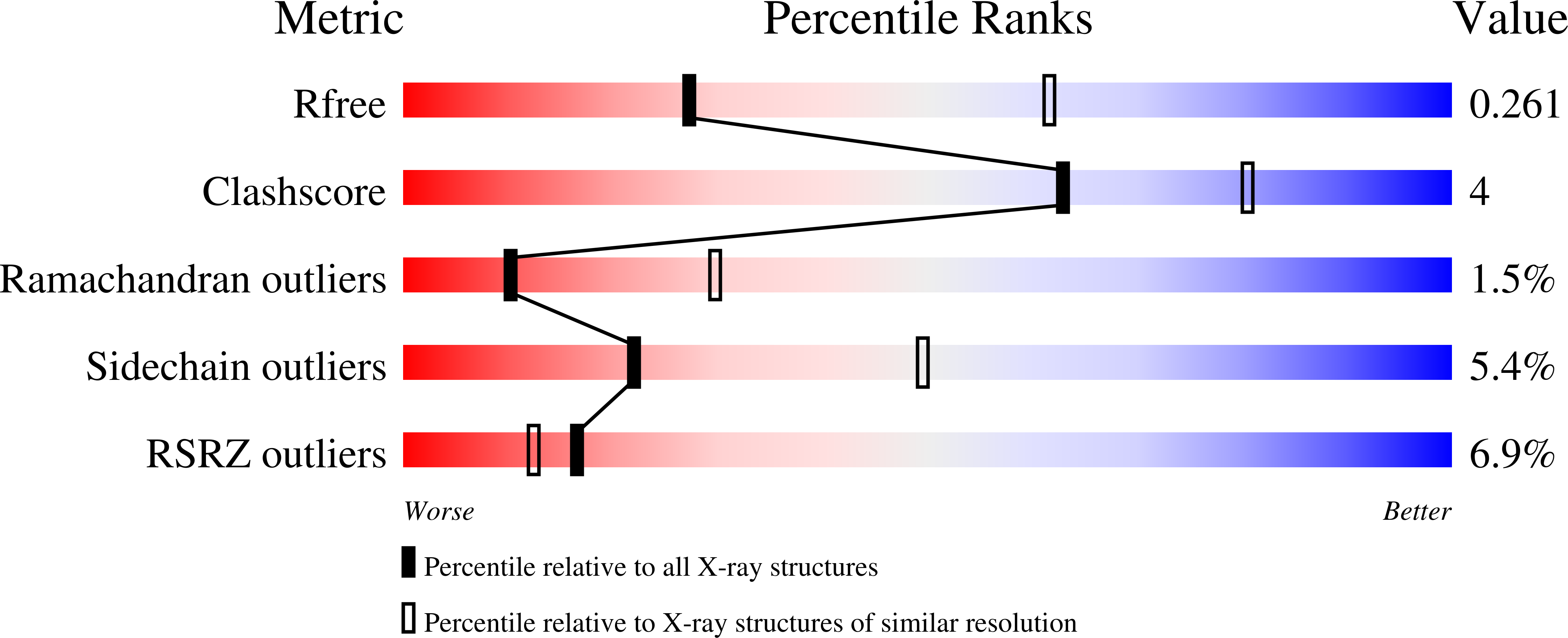
Engineering of an isolated p110 alpha subunit of PI3K alpha permits crystallization and provides a platform for structure-based drug design.
Chen, P., Deng, Y.L., Bergqvist, S., Falk, M.D., Liu, W., Timofeevski, S., Brooun, A.(2014) Protein Sci 23: 1332-1340
- PubMed: 25043846
- DOI: https://doi.org/10.1002/pro.2517
- Primary Citation of Related Structures:
4TUU, 4TV3 - PubMed Abstract:
PI3K¦Á remains an attractive target for the development of anticancer targeted therapy. A number of p110¦Á crystal structures in complex with the nSH2-iSH2 fragment of p85 regulatory subunit have been reported, including a few small molecule co-crystal structures, but the utilization of this crystal form is limited by low diffraction resolution and a crystal packing artifact that partially blocks the ATP binding site. Taking advantage of recent data on the functional characterization of the lipid binding properties of p110¦Á, we designed a set of novel constructs allowing production of isolated stable p110¦Á subunit missing the Adapter Binding Domain and lacking or featuring a modified C-terminal lipid binding motif. While this protein is not catalytically competent to phosphorylate its substrate PIP2, it retains ligand binding properties as indicated by direct binding studies with a pan-PI3K¦Á inhibitor. Additionally, we determined apo and PF-04691502 bound crystal structures of the p110¦Á (105-1048) subunit at 2.65 and 2.85 ?, respectively. Comparison of isolated p110¦Á(105-1048) with the p110¦Á/p85 complex reveals a high degree of structural similarity, which validates suitability of this catalytically inactive p110¦Á for iterative SBDD. Importantly, this crystal form of p110¦Á readily accommodates the binding of noncovalent inhibitor by means of a fully accessible ATP site. The strategy presented here can be also applied to structural studies of other members of PI3KIA family.
Organizational Affiliation:
Oncology Structural Biology, Worldwide Research and Development, Pfizer Inc., San Diego, California, 92121.