RCSB PDB Help
General Help
Computed Structure Models and RCSB.org
Video: What is in a Computed Structure Model?
Introduction
Protein Data Bank and RCSB.org
The Protein Data Bank (PDB) archives experimentally determined three dimensional (3D) structural data of biological macromolecules. In addition to providing access to 3D structural data, all members of the worldwide PDB (wwPDB) offer tools to query the archive, and then organize, visualize, and analyze groups of structures to learn about any topic of interest. The Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), website (RCSB.org) integrates information about properties and functions of proteins from a variety of publicly available bioinformatics data resources - e.g., information about gene sequences, mutations, disease correlations, small molecule (drug) binding affinities. Mapping information integrated from these resources to 3D structural data can provide insights beyond what is available from the molecular structures alone. Users can access the structural and integrated information from the RCSB.org website for new perspectives about the topic of interest and ask new questions.
Expanding the limits of PDB
Although the PDB archive continues to rapidly grow in size and complexity, there are many millions of proteins whose structures have not yet been solved. For the past few decades researchers have been developing a variety of approaches for reliably predicting computed structure models of proteins. In 2020, two different projects [AlphaFold2 (Jumper, J. et al., 2021, Varadi M. et al., 2022) and RoseTTAFold (Baek et al., 2021)] used artificial intelligence (AI) and machine learning (ML) to successfully predict protein structures from their sequences. The approaches utilize knowledge of protein structures from the PDB, and vast amounts of protein sequencing data to compute these models.
Access to reliable computed structure models (CSMs) has created new opportunities for molecular explorations and analysis. When experimental structures of the protein or complexes being studied are not available, CSMs can provide a great alternative and/or an initial model for data analysis, and hypothesis development. To make it easier for users to query, organize, visualize, analyze and compare experimental and predicted structures alongside each other, RCSB PDB has integrated CSMs from a few specific resources.
Experimental Structures and CSM at RCSB.org
Access to both experimental structure and CSMs on a topic of interest offers users insights and choice. When exploring structure-function relationships, experimental structures of a protein or parts of it are likely to be more accurate and have a higher level of confidence compared to CSMs. Yet CSMs can provide great starting models for the millions of proteins and their complexes whose structures have not been experimentally determined. It should be noted that the quality of CSMs may not be uniform - parts of the CSM that are computed based on existing PDB structures and various other experimental data are likely to be more accurate and predicted with a higher level of confidence. Learn more about the quality and confidence of experimental structures and CSMs. As with experimental structures in the PDB, when using CSMs the accuracy and confidence of the 3D models should be considered in structure analysis and hypothesis development.
Documentation
What CSMs are available?
The computed structure models from the following providers are integrated with RCSB.org:
- AlphaFold Protein Structure Database. These models are state-of-the-art protein structure predictions based on amino-acid sequences, using an AI system called AlphaFold2 (Jumper et al., 2021, Varadi M. et al., 2022). Here, we have included ~1,000,000 of these models at RCSB.org, using the pre-packaged collections of models made publicly available on https://ftp.ebi.ac.uk/pub/databases/alphafold/, which encompass four main groups of models (as listed on https://alphafold.ebi.ac.uk/download):
- Model organism proteomes: Protein structure models from >40 different model organisms e.g., Arabidopsis, E. coli, fruit fly, human, soybean, and zebrafish.
- Global health proteomes: Protein structure models from various disease-causing organisms, e.g., H. pylori, K. pneumoniae, M. tuberculosis, and P. falciparum.
- Swiss-Prot sequences (the manually curated set of UniProt sequences).
- MANE (Matched Annotation from NCBI and EMBL-EBI) select sequences.
- ModelArchive: This database hosts user-submitted predictions of protein structures generated using a variety of approaches, e.g., homology modeling, ab initio, and deep learning techniques. The following CSM datasets from the ModelArchive have been integrated with RCSB.org:
- ma-bak-cepc: Core eukaryotic protein complexes produced by the Baker lab using a combination of RoseTTAFold (Baek et al., 2021) and AlphaFold2 (Humphreys et al., 2021). Find these 3D structures.
- ma-coffe-slac: Freshwater sponge proteins (modeled with ColabFold). Find these 3D structures.
- ma-asfv-asfvg: African swine fever virus proteins (modeled with AlphaFold). Find these 3D structures.
- ma-ornl-sphdiv: Structural models of the Sphagnum divinum proteome modeled using AlphaFold2 (https://ieeexplore.ieee.org/document/9835405). Find these 3D structures.
- ma-t3vr3: Hetero-dimer set of proteins from cancer interactome modeled using AlphaFold (https://pubmed.ncbi.nlm.nih.gov/36261849/). Find these 3D structures.
How can you access the CSMs?
The following approaches are available to identify experimental structures and CSMs in RCSB.org and query for them.
Identifying Type of 3D Structure
Specific icons (dark blue flask icon for experimental structures and cyan colored computer icon for CSMs) are now used throughout the website to quickly and easily identify the source of 3D models selected for visualization and/or analysis.
Querying for Structures in RCSB.org
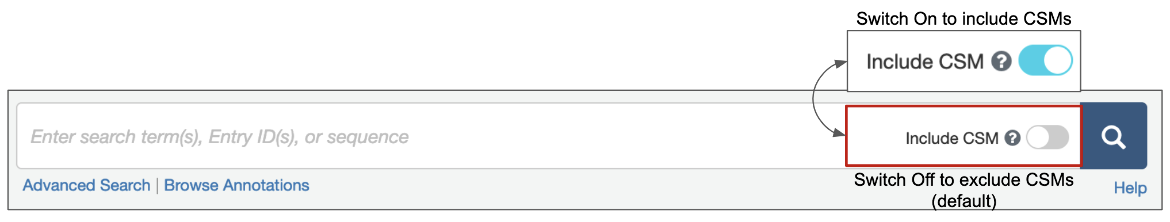
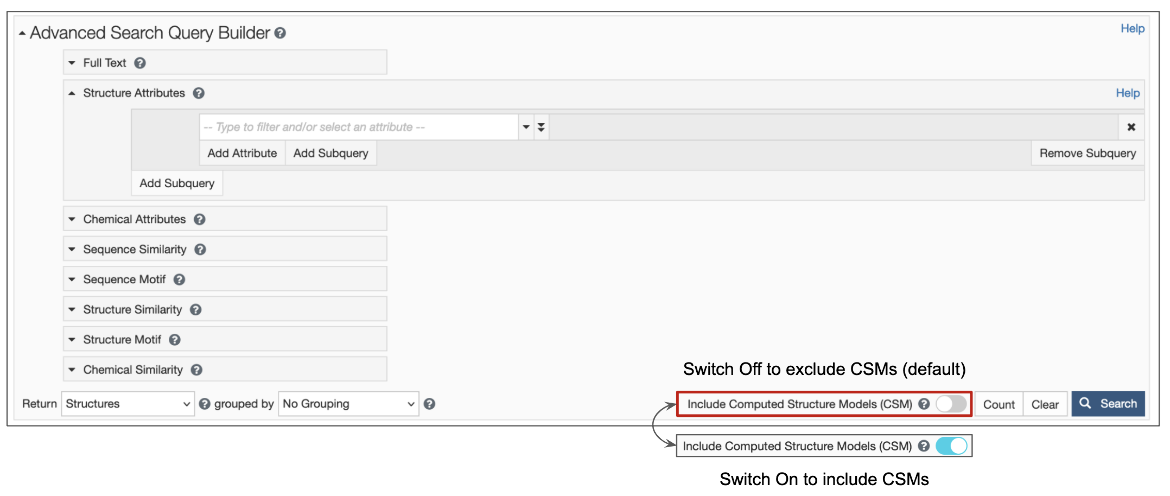
1. Options are available for structure queries to include CSMs alongside experimental structures (from the PDB) in the search results. When the toggle switch in the top search box is turned 'on' (i.e., is cyan-colored, as shown in Figure 1), CSMs are included in the search. Learn more about including/excluding CSMs in basic search. The Advanced Search Query builder also has a similar toggle switch to include CSMs (Figure 2).
|
| Figure 2: Advanced Search Query Builder options available form the RCSB.org home page |
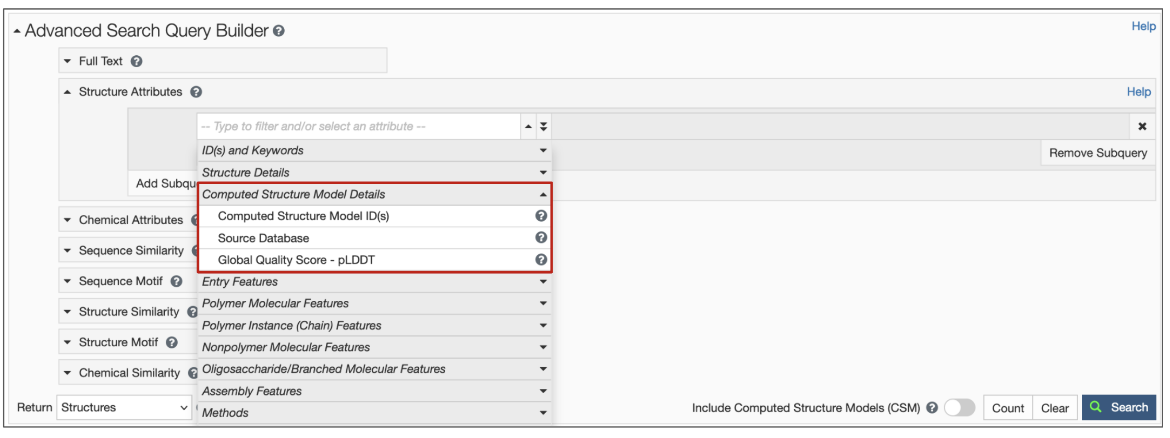
2. New structure attributes for CSMs have been added to search options so that specific queries based on source database and confidence level can be made (Figure 3). Learn more about the Computed Structure Model Attributes.
|
| Figure 3: Structure Attributes (properties) available to search for CSMs using the Advanced Search Query Builder. |
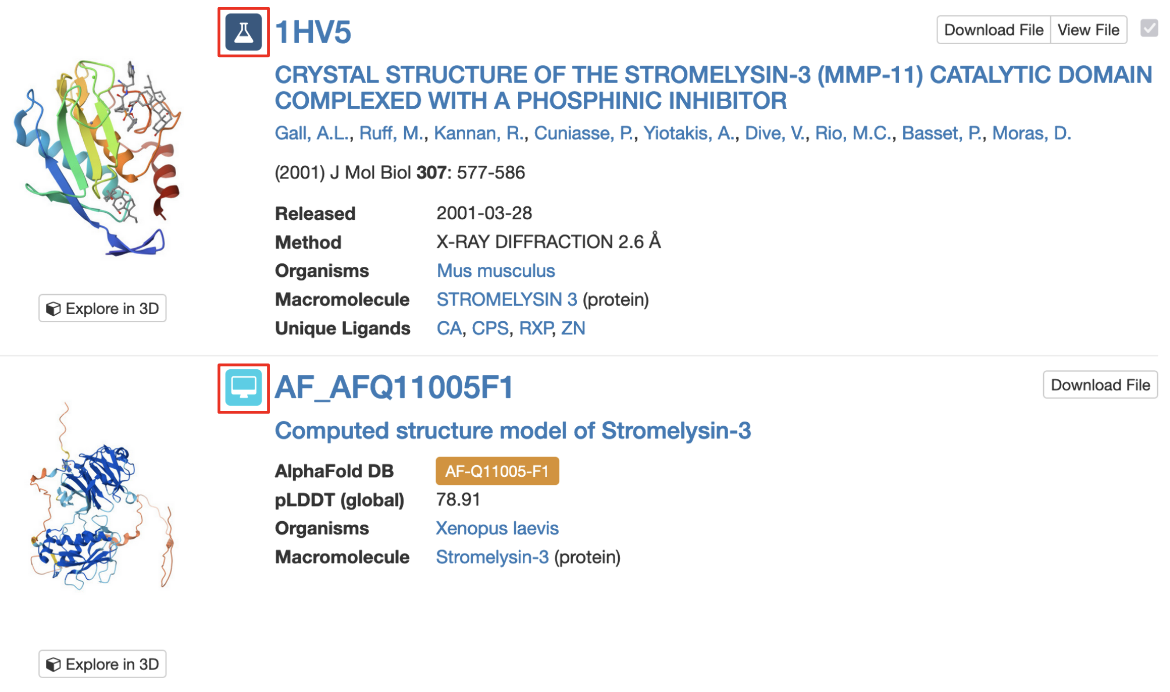
3. Query results include icons to indicate whether matched structure(s) are experimental models or CSMs (Figure 4).
|
| Figure 4: Part of the Search Results page showing an experimental structure and a CSM, each marked with their respective icons (highlighted with a red outlined box). |
Note: Each CSM is assigned a specific ID in its source database and a prefix indicates the source of the model (¡°AF¡± for AlphaFold DB, "MA" for ModelArchive). AlphaFold DB identifiers are then followed by the UniProt accession number for the protein and by the fragment number (usually ¡°F1¡±). However, in order to enable compatibility of the IDs with many of our services, including all of our APIs and visualization tools, we identify CSMs on RCSB.org using a modified version of the ID. This ID is used on the structure summary page, in searching for structures, in the search results page, and in various tools for 3D structure visualization and analysis. For example, for the AlphaFold structure AF-B3EWR1-F1, the RCSB.org assigned CSM ID is AF_AFB3EWR1F1 and is used in the query results page as shown in Figure 4.
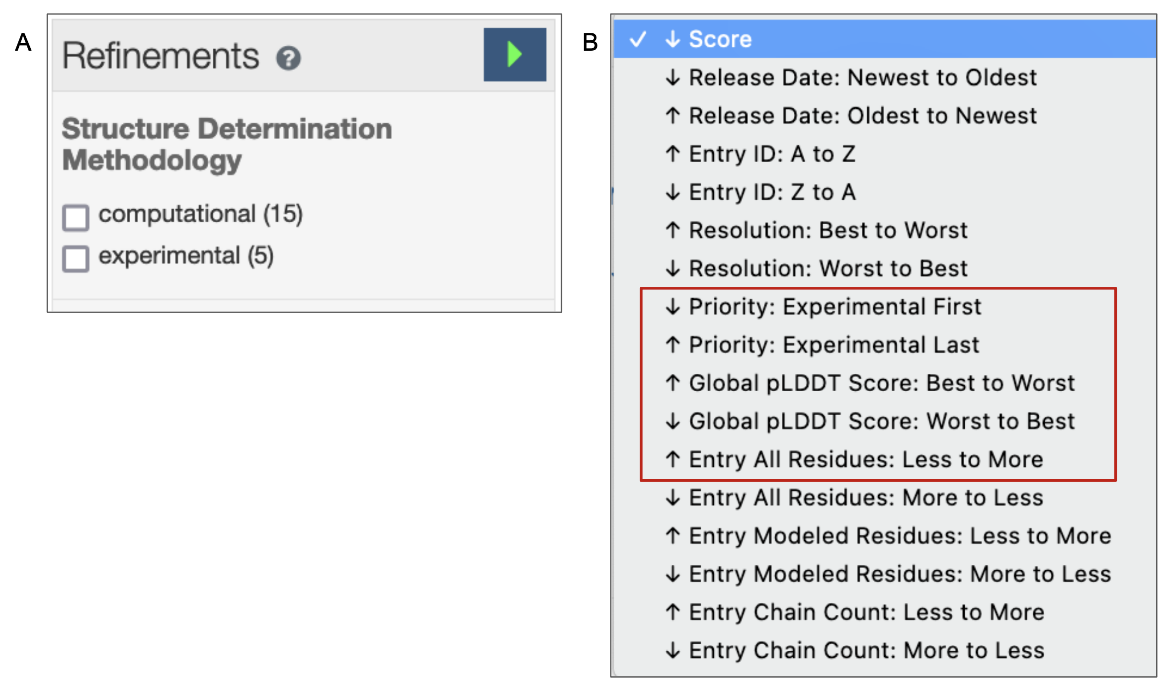
4. The default order of the search results is based on a relevancy score for the query criteria. The Refinements menu on the left of the query results page offers options to view only experimental structures or only CSMs in the search results (see Figure 5A). Learn more about the query results page and refinements. The order of the search results may also be changed according to a few options - e.g., view CSMs in the results first or last; order the CSMs by pLDDT scores (see Figure 5B).
What can you do with the CSMs at RCSB.org?
You can search for, explore, visualize, analyze, and compare experimental structures and CSMs at RCSB.org.
- Search for 3D structures (experimental structures and CSMs) using a variety of search services, including searches based on attributes, sequences, sequence motifs, structures, and structure motifs.
- View individual CSM structure summary pages that provide a quick overview of quality based on confidence levels defined by pLDDT scores. Learn more here.
- Each CSM structure summary page has options to visualize and analyze its 3D structure in a manner identical to that provided for experimental structures in the PDB.
- Download the 3D coordinates of any specific CSM structure from either the query results page or structure summary page. Note that requests to download batches of multiple CSM structures should be directed to the relevant model source database.
- Group CSM structures together with experimental structures to generate group views for comparison and analysis.
- Perform comparative analyses of protein 3D structures to find similarities within a set of CSMs or between CSM and PDB structures using the pairwise structure alignment tool.
Models and Assembly
The CSMs available from the RCSB.org make no claims about predicting higher order oligomeric assemblies. However, to include CSMs in structure based query and analysis (e.g., Find similar assembly, Structure search, Structure motif search), the model coordinates of CSMs are also included as Assemblies - i.e., for CSMs the Model and Assembly coordinates are identical.
Examples
- Query for proteins (including CSMs) from the Mediterranean mussel (Mytilus galloprovincialis).
- Structure summary page for the Histone-4 protein of Mytilus galloprovincialis (from AlphaFoldDB).
- Query for high-quality (pLDDT > 90) computed structure models of human proteins.
- Query for 3D structures of myoglobin grouped by 30% sequence identity and displayed as groups.
- Query for mouse CSMs that do not have a corresponding experimental structure.
For Advanced Users
Advanced users who wish to access CSMs programmatically can use web-services:
- to run searches using RCSB PDB Search API. See example
- to query data using RCSB PDB Data API. See example
- to retrieve sequence alignments for CSMs using RCSB PDB 1D Coordinates API. See example
Further details on Web Services and APIs are available at Web Services Overview.
References
- Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., Wang, J., Cong, Q., Kinch, L. N., Schaeffer, R. D., Mill¨¢n, C., Park, H., Adams, C., Glassman, C. R., DeGiovanni, A., Pereira, J. H., Rodrigues, A. V., van Dijk, A. A., Ebrecht, A. C., Opperman, D. J., ¡ Baker, D. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science, 373, 871¨C876. https://doi.org/10.1126/science.abj8754
- Jumper, J., Evans, R., Pritzel, A. et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596, 583¨C589 . https://doi.org/10.1038/s41586-021-03819-2
- Humphreys, I. R., Pei, J., Baek, M., Krishnakumar, A., Anishchenko, I., Ovchinnikov, S., Zhang, J., Ness, T. J., Banjade, S., Bagde, S. R., Stancheva, V. G., Li, X. H., Liu, K., Zheng, Z., Barrero, D. J., Roy, U., Kuper, J., Fern¨¢ndez, I. S., Szakal, B., Branzei, D., ¡ Baker, D. (2021). Computed structures of core eukaryotic protein complexes. Science, 374(6573), eabm4805. https://doi.org/10.1126/science.abm4805
- Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., Yuan, D., Stroe, O., Wood, G., Laydon, A., ?¨ªdek, A., Green, T., Tunyasuvunakool, K., Petersen, S., Jumper, J., Clancy, E., Green, R., Vora, A., Lutfi, M., Figurnov, M., Cowie, A., Hobbs, N., Kohli, P., Kleywegt, G., Birney, E., Hassabis, D., Velankar, S. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models, Nucleic Acids Research, 50, D439¨CD444, https://doi.org/10.1093/nar/gkab1061