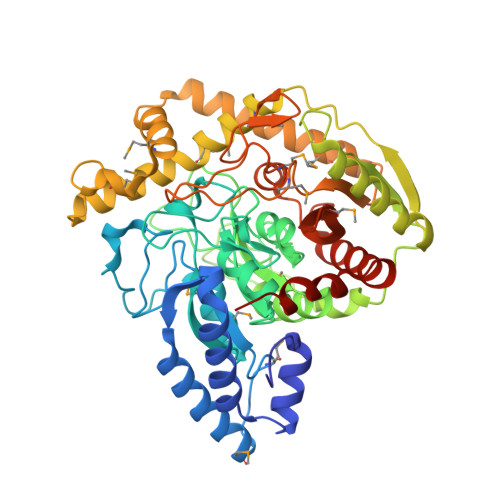
Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Lee, S., Park, E.-H., Ko, H.-J., Bang, W.-G., Kim, H.Y., Kim, K.H., Choi, I.-G.(2015) Biochem Biophys Res Commun 467: 268-274
- PubMed: 26454172
- DOI: https://doi.org/10.1016/j.bbrc.2015.09.177
- Primary Citation of Related Structures:
4YJ6, 4YJI - PubMed Abstract:
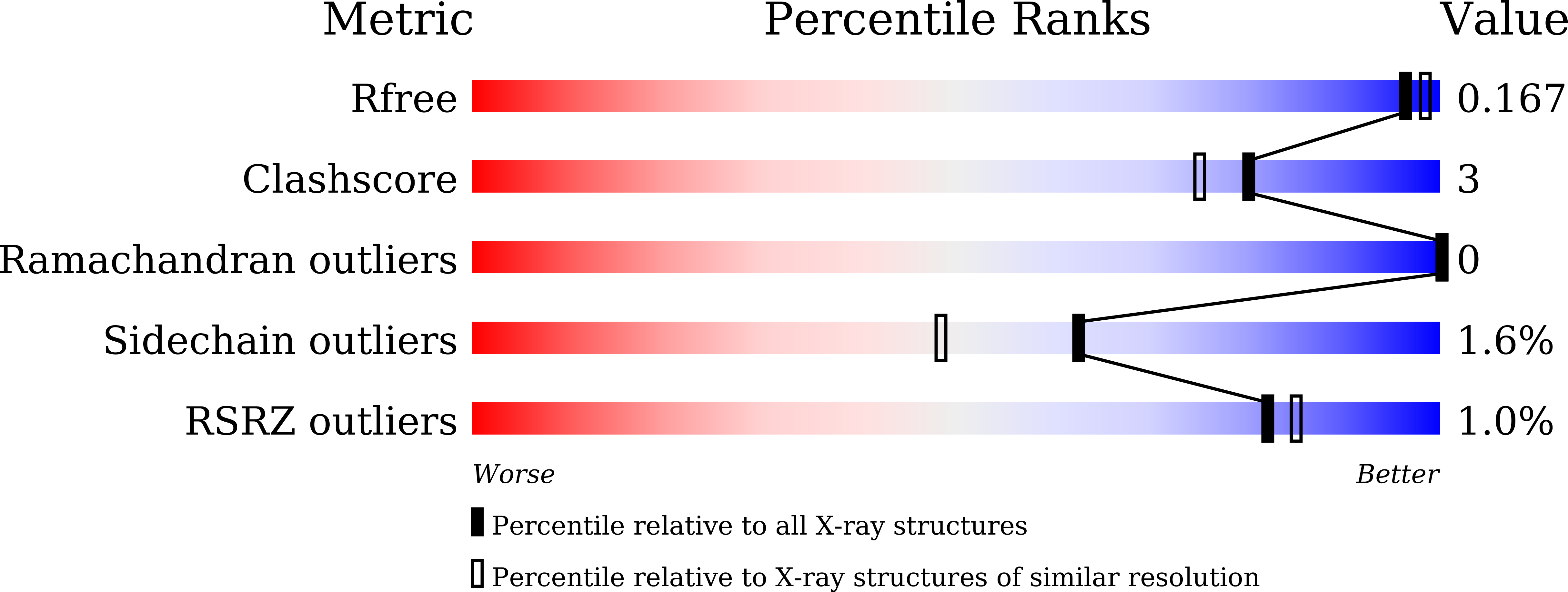
The atomic structure of a bacterial aryl acylamidase (EC 3.5.1.13; AAA) is reported and structural features are investigated to better understand the catalytic profile of this enzyme. Structures of AAA were determined in its native form and in complex with the analgesic acetanilide, p-acetaminophenol, at 1.70?? and 1.73?? resolutions, respectively. The overall structural fold of AAA was identified as an ¦Á/¦Â fold class, exhibiting an open twisted ¦Â-sheet core surrounded by ¦Á-helices. The asymmetric unit contains one AAA molecule and the monomeric form is functionally active. The core structure enclosing the signature sequence region, including the canonical Ser-cisSer-Lys catalytic triad, is conserved in all members of the Amidase Signature enzyme family. The structure of AAA in a complex with its ligand reveals a unique organization in the substrate-binding pocket. The binding pocket consists of two loops (loop1 and loop2) in the amidase signature sequence and one helix (¦Á10) in the non-amidase signature sequence. We identified two residues (Tyr(136) and Thr(330)) that interact with the ligand via water molecules, and a hydrogen-bonding network that explains the catalytic affinity over various aryl acyl compounds. The optimum activity of AAA at pH?>?10 suggests that the reaction mechanism employs Lys(84) as the catalytic base to polarize the Ser(187) nucleophile in the catalytic triad.
Organizational Affiliation:
Department of Biotechnology, College of Life Sciences and Biotechnology, Korea University, Anam-Dong, Seoungbuk-Gu, Seoul, 136-713, South Korea.