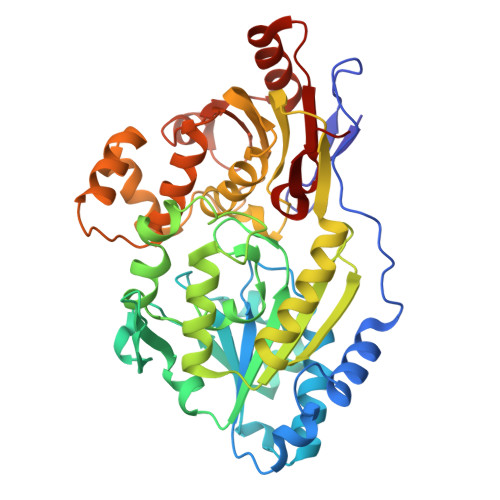
Structural Basis for Catalysis and Substrate Specificity of a LarA Racemase with a Broad Substrate Spectrum.
Gatreddi, S., Urdiain-Arraiza, J., Desguin, B., Hausinger, R.P., Hu, J.(2024) bioRxiv
- PubMed: 39651260
- DOI: https://doi.org/10.1101/2024.11.28.625916
- Primary Citation of Related Structures:
9EIA, 9EID, 9EIF - PubMed Abstract:
The LarA family consists of diverse racemases/epimerases that interconvert the diastereomers of a variety of ¦Á-hydroxyacids by using a nickel-pincer nucleotide (NPN) cofactor. The hidden redox reaction catalyzed by the NPN cofactor makes LarA enzymes attractive engineering targets for applications. However, how a LarA enzyme binds its natural substrate and recognizes different ¦Á-hydroxyacids has not been elucidated. Here, we report three high-resolution structures of the enzyme-substrate complexes of a broad-spectrum LarA enzyme from Isosphaera pallida (LarA Ip ). The substrate binding mode reveals an optimal orientation and distance between the hydride donor and acceptor, strongly supporting the proposed proton-coupled hydride transfer mechanism. The experimentally solved structures, together with the structural models of other LarA enzymes, allow us to identify the residues/structural elements critically involved in the interactions with different ¦Á-hydroxyacid substrates. Collectively, this work provides a critical structural basis for catalysis and substrate recognition of the diverse enzymes in the LarA family, thus building a foundation for enzyme engineering.