News
New version of PDB-IHM validation reports now include assessments of crosslinking mass spectrometry-based structure models
03/10 
 New version of PDB-IHM validation reports now include assessments of crosslinking mass spectrometry-based structure models
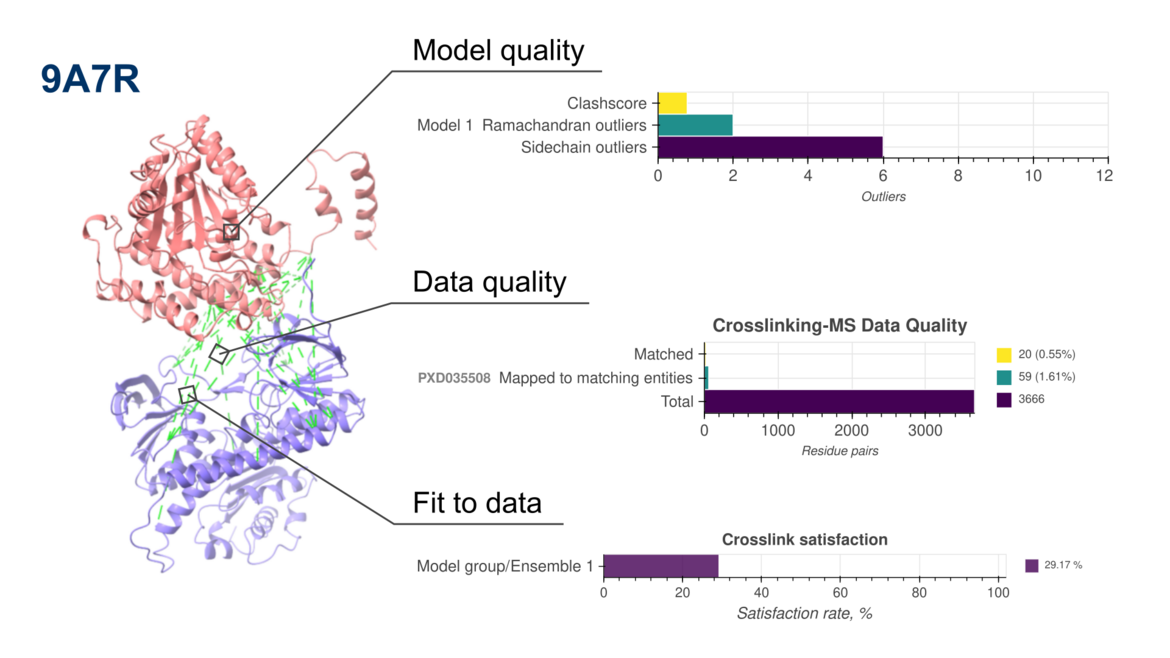
New version of PDB-IHM validation reports now include assessments of crosslinking mass spectrometry-based structure modelsWe are pleased to announce the release of updated validation reports (version 2) for structures determined using integrative and hybrid methods (IHM) archived in the PDB-IHM branch of the PDB archive. These reports are available in the archive and on the PDB-IHM website. IHM Validation software v2 is now integrated into the PDB-IHM deposition and curation system to support validation of new depositions. For detailed information on IHM Validation and v2 updates, please refer to the help page and user guide. The software is also available for standalone use via GitHub.
We thank the wwPDB IHM Task Force members and the broader structural biology community for supporting the development of validation methods for integrative structures.
What’s New in Version 2?
The latest update extends IHM validation to structures derived from Chemical Crosslinking Mass Spectrometry (Crosslinking-MS) data, in addition to the previously supported Small Angle Scattering (SAS) data. This milestone is the result of close collaboration among the Crosslinking-MS community, PRIDE repository, and the PDB-IHM team.
To support this advancement, we have developed interoperability mechanisms between PDB-IHM and PRIDE, enabling validation of integrative structures based on Crosslinking-MS data. Additionally, PRIDE Crosslinking now supports submission of complete Crosslinking-MS datasets following the mzIdentML 1.2.0 community data standard.
Furthermore, IHM Validation software v2 uses MolProbity version 4.5.2 and the PrISM software for precision analysis.
Next Steps for Depositors
We encourage depositors working with Crosslinking-MS-based integrative structures to:
- Submit complete datasets to PRIDE Crosslinking following the provided guidelines.
- Cross-reference PRIDE Crosslinking datasets in PDB-IHM depositions to enable robust validation.














