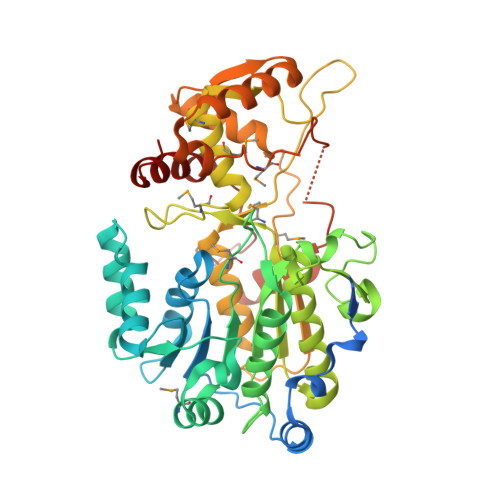
Nonprocessive [2 + 2]e- off-loading reductase domains from mycobacterial nonribosomal peptide synthetases.
Chhabra, A., Haque, A.S., Pal, R.K., Goyal, A., Rai, R., Joshi, S., Panjikar, S., Pasha, S., Sankaranarayanan, R., Gokhale, R.S.(2012) Proc Natl Acad Sci U S A 109: 5681-5686
- PubMed: 22451903
- DOI: https://doi.org/10.1073/pnas.1118680109
- Primary Citation of Related Structures:
4DQV - PubMed Abstract:
In mycobacteria, polyketide synthases and nonribosomal peptide synthetases (NRPSs) produce complex lipidic metabolites by using a thio-template mechanism of catalysis. In this study, we demonstrate that off-loading reductase (R) domain of mycobacterial NRPSs performs two consecutive [2?+?2]e(-) reductions to release thioester-bound lipopeptides as corresponding alcohols, using a nonprocessive mechanism of catalysis. The first crystal structure of an R domain from Mycobacterium tuberculosis NRPS provides strong support to this mechanistic model and suggests that the displacement of intermediate would be required for cofactor recycling. We show that 4e(-) reductases produce alcohols through a committed aldehyde intermediate, and the reduction of this intermediate is at least 10 times more efficient than the thioester-substrate. Structural and biochemical studies also provide evidence for the conformational changes associated with the reductive cycle. Further, we show that the large substrate-binding pocket with a hydrophobic platform accounts for the remarkable substrate promiscuity of these domains. Our studies present an elegant example of the recruitment of a canonical short-chain dehydrogenase/reductase family member as an off-loading domain in the context of assembly-line enzymology.
Organizational Affiliation:
National Institute of Immunology, Aruna Asaf Ali Marg, New Delhi 110067, India.