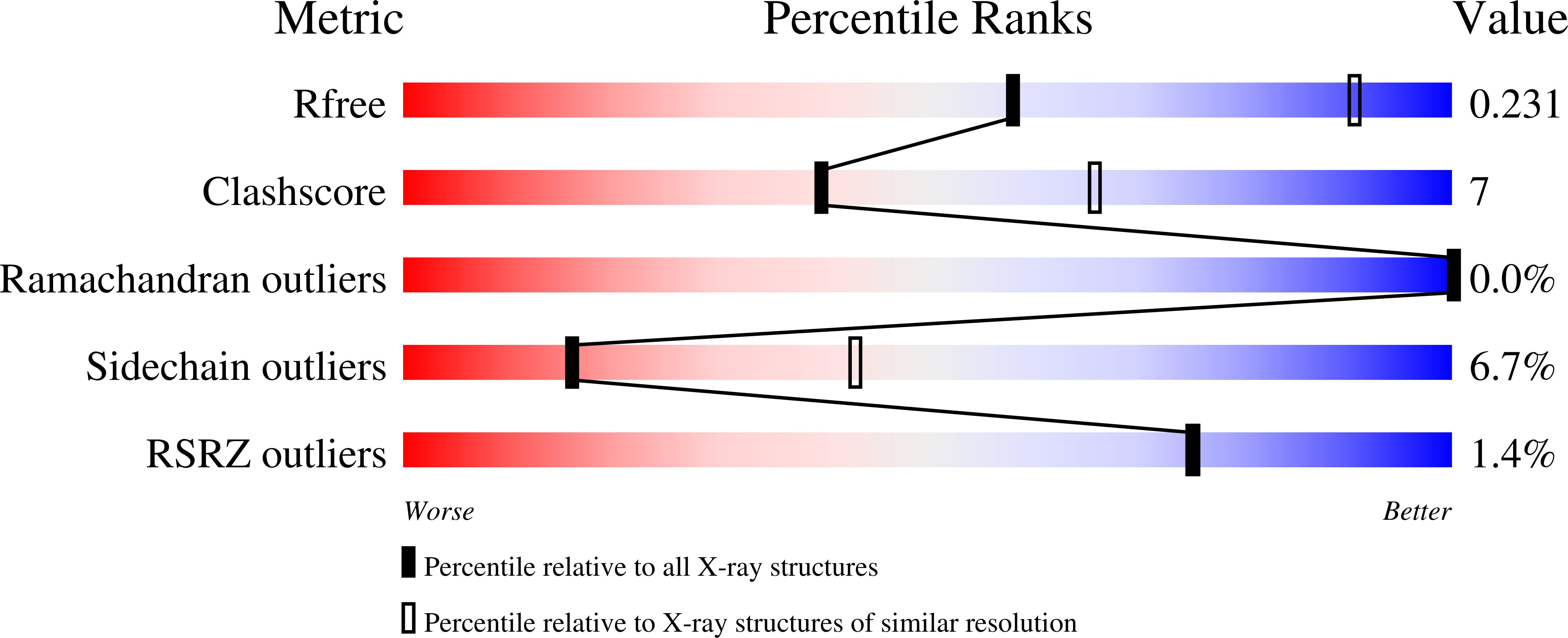
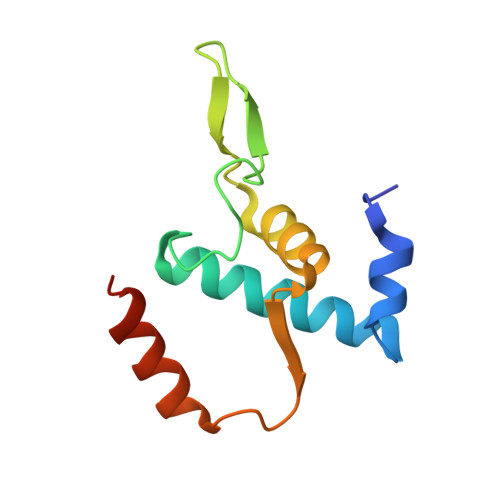
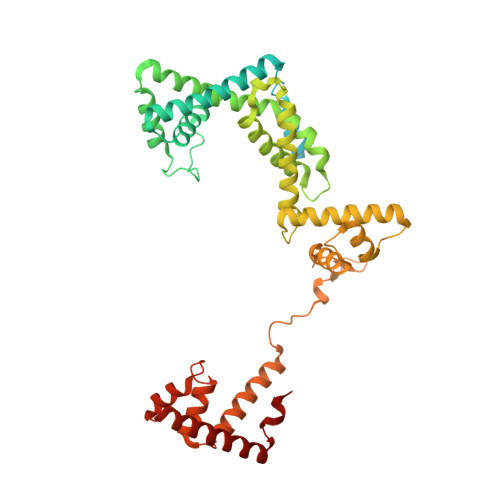
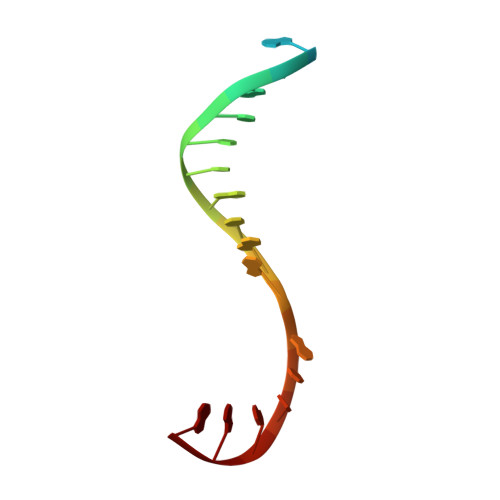
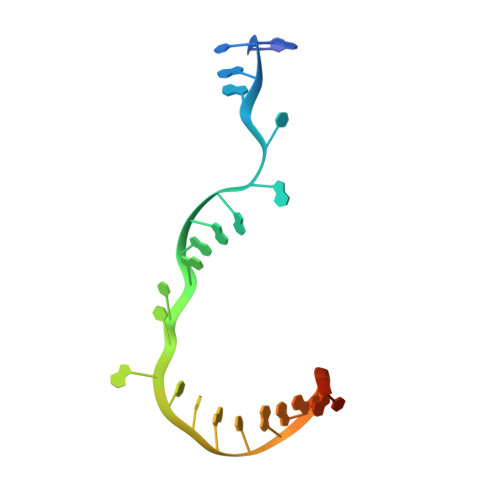
Structural basis of transcription initiation.
Zhang, Y., Feng, Y., Chatterjee, S., Tuske, S., Ho, M.X., Arnold, E., Ebright, R.H.(2012) Science 338: 1076-1080
- PubMed: 23086998
- DOI: https://doi.org/10.1126/science.1227786
- Primary Citation of Related Structures:
4G7H, 4G7O, 4G7Z - PubMed Abstract:
During transcription initiation, RNA polymerase (RNAP) binds and unwinds promoter DNA to form an RNAP-promoter open complex. We have determined crystal structures at 2.9 and 3.0 ? resolution of functional transcription initiation complexes comprising Thermus thermophilus RNA polymerase, ¦̉(A), and a promoter DNA fragment corresponding to the transcription bubble and downstream double-stranded DNA of the RNAP-promoter open complex. The structures show that ¦̉ recognizes the -10 element and discriminator element through interactions that include the unstacking and insertion into pockets of three DNA bases and that RNAP recognizes the -4/+2 region through interactions that include the unstacking and insertion into a pocket of the +2 base. The structures further show that interactions between ¦̉ and template-strand single-stranded DNA (ssDNA) preorganize template-strand ssDNA to engage the RNAP active center.
Organizational Affiliation:
Howard Hughes Medical Institute, Waksman Institute, and Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854, USA.