Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer.
Pica, A., Russo Krauss, I., Merlino, A., Nagatoishi, S., Sugimoto, N., Sica, F.(2013) FEBS J 280: 6581-6588
- PubMed: 24128303
- DOI: https://doi.org/10.1111/febs.12561
- Primary Citation of Related Structures:
4LZ1, 4LZ4 - PubMed Abstract:
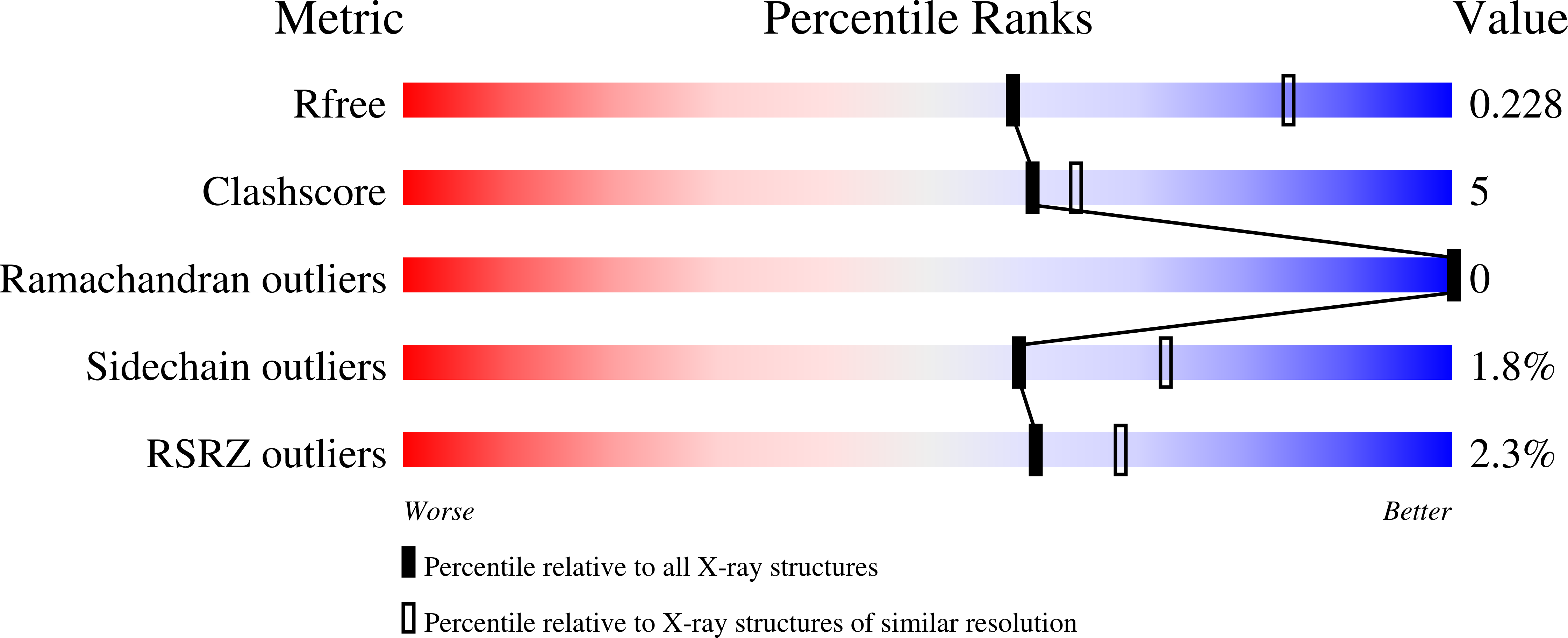
Thrombin plays a pivotal role in the coagulation cascade; therefore, it represents a primary target in the treatment of several blood diseases. The 15-mer DNA oligonucleotide 5'-GGTTGGTGTGGTTGG-3', known as thrombin binding aptamer (TBA), is a highly potent inhibitor of the enzyme. TBA folds as an antiparallel chair-like G-quadruplex structure, with two G-tetrads surrounded by two TT loops on one side and a TGT loop on the opposite side. Previous crystallographic studies have shown that TBA binds thrombin exosite I by its TT loops, T3T4 and T12T13. In order to get a better understanding of the thrombin-TBA interaction, we have undertaken a crystallographic characterization of the complexes between thrombin and two TBA mutants, TBA¦¤T3 and TBA¦¤T12, which lack the nucleobase of T3 and T12, respectively. The structural details of the two complexes show that exosite I is actually split into two regions, which contribute differently to TBA recognition. These results provide the basis for a more rational design of new aptamers with improved therapeutic action.
Organizational Affiliation:
Department of Chemical Sciences, University of Naples Federico II, Italy.