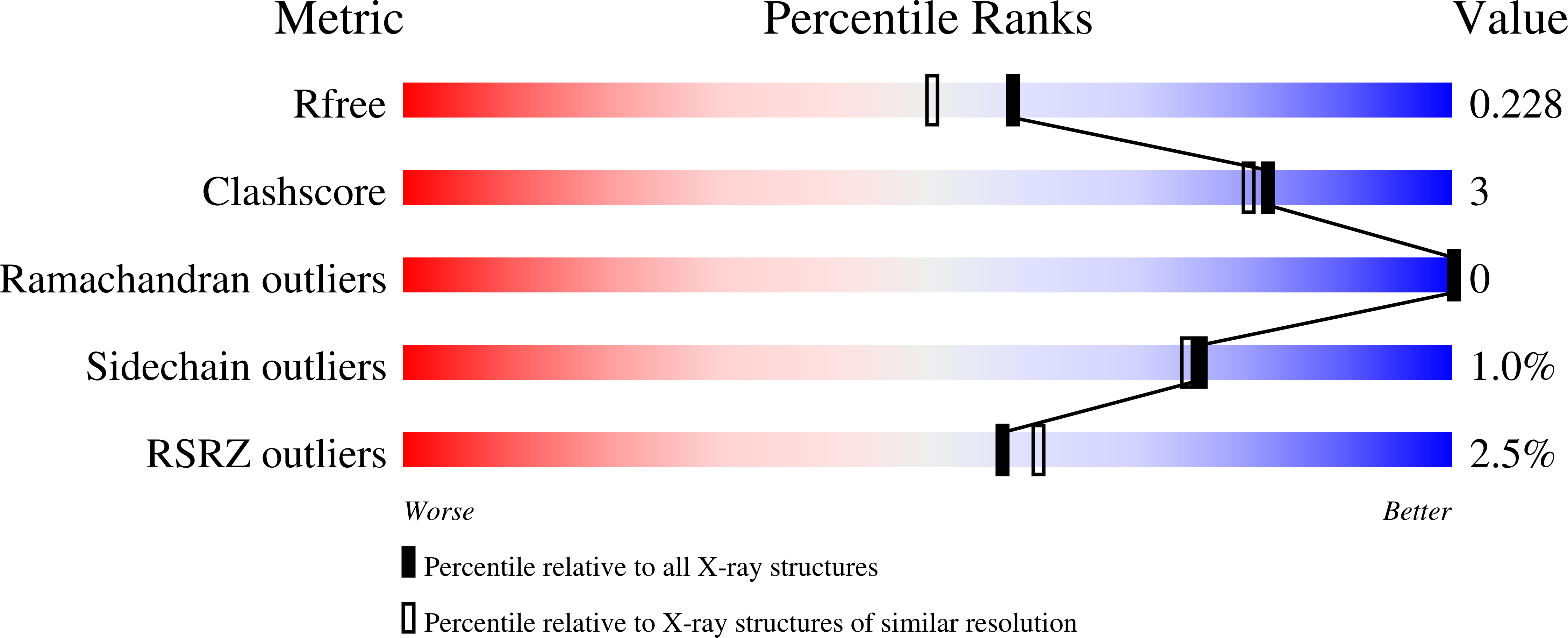
Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Holden, J.K., Dejam, D., Lewis, M.C., Huang, H., Kang, S., Jing, Q., Xue, F., Silverman, R.B., Poulos, T.L.(2015) Biochemistry 54: 4075
- PubMed: 26062720
- DOI: https://doi.org/10.1021/acs.biochem.5b00431
- Primary Citation of Related Structures:
4UG5, 4UG6, 4UG7, 4UG8, 4UG9, 4UGA, 4UGB, 4UGC, 4UGD, 4UGE, 4UGF, 4UGG, 4UGH, 4UGI, 4UGJ, 4UGK, 4UGL, 4UGM, 4UGN, 4UGO, 4UGP, 4UGQ, 4UGR, 4UGS, 4UGT, 4UGU, 4UGV, 4UGW, 4UGX, 4UGY - PubMed Abstract:
Nitric oxide generated by bacterial nitric oxide synthase (NOS) increases the susceptibility of Gram-positive pathogens Staphylococcus aureus and Bacillus anthracis to oxidative stress, including antibiotic-induced oxidative stress. Not surprisingly, NOS inhibitors also improve the effectiveness of antimicrobials. Development of potent and selective bacterial NOS inhibitors is complicated by the high active site sequence and structural conservation shared with the mammalian NOS isoforms. To exploit bacterial NOS for the development of new therapeutics, recognition of alternative NOS surfaces and pharmacophores suitable for drug binding is required. Here, we report on a wide number of inhibitor-bound bacterial NOS crystal structures to identify several compounds that interact with surfaces unique to the bacterial NOS. Although binding studies indicate that these inhibitors weakly interact with the NOS active site, many of the inhibitors reported here provide a revised structural framework for the development of new antimicrobials that target bacterial NOS. In addition, mutagenesis studies reveal several key residues that unlock access to bacterial NOS surfaces that could provide the selectivity required to develop potent bacterial NOS inhibitors.
Organizational Affiliation:
Departments of ?Molecular Biology and Biochemistry, ?Pharmaceutical Sciences, and ¡́Chemistry, University of California, Irvine, California 92697-3900, United States.