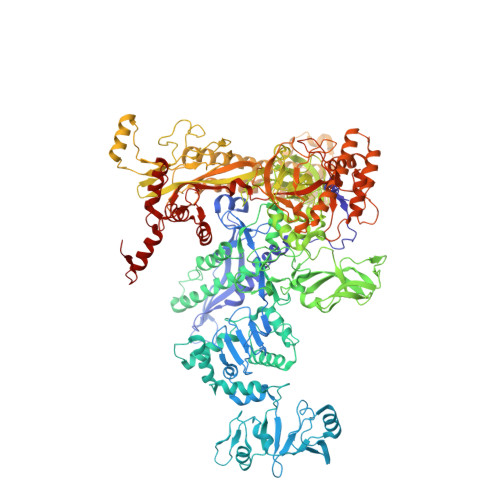
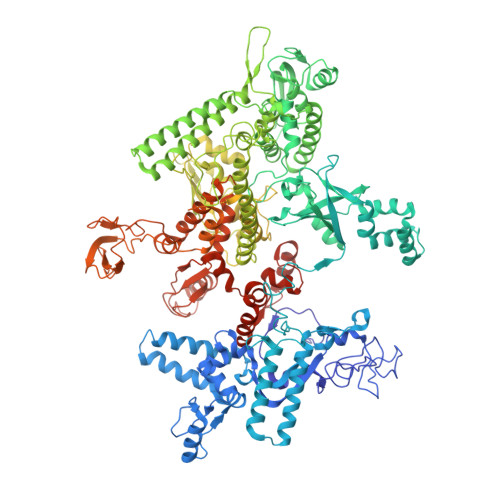
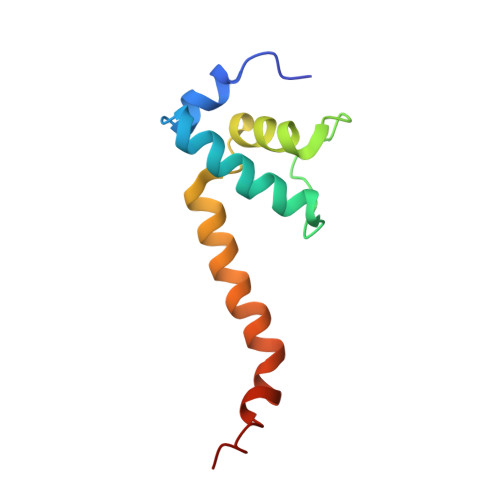
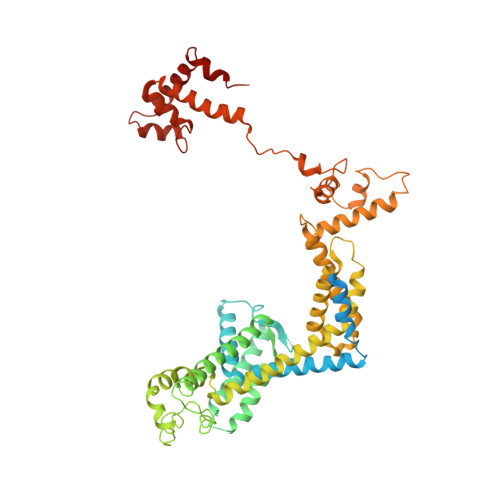
X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme
Murakami, K.S.(2013) J Biol Chem 288: 9126-9134
- PubMed: 23389035
- DOI: https://doi.org/10.1074/jbc.M112.430900
- Primary Citation of Related Structures:
4YG2 - PubMed Abstract:
Escherichia coli RNA polymerase (RNAP) is the most studied bacterial RNAP and has been used as the model RNAP for screening and evaluating potential RNAP-targeting antibiotics. However, the x-ray crystal structure of E. coli RNAP has been limited to individual domains. Here, I report the x-ray structure of the E. coli RNAP 考(70) holoenzyme, which shows 考 region 1.1 (考1.1) and the 汐 subunit C-terminal domain for the first time in the context of an intact RNAP. 考1.1 is positioned at the RNAP DNA-binding channel and completely blocks DNA entry to the RNAP active site. The structure reveals that 考1.1 contains a basic patch on its surface, which may play an important role in DNA interaction to facilitate open promoter complex formation. The 汐 subunit C-terminal domain is positioned next to 考 domain 4 with a fully stretched linker between the N- and C-terminal domains. E. coli RNAP crystals can be prepared from a convenient overexpression system, allowing further structural studies of bacterial RNAP mutants, including functionally deficient and antibiotic-resistant RNAPs.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Center for RNA Molecular Biology, Pennsylvania State University, University Park, PA 16802, USA. kum14@psu.edu