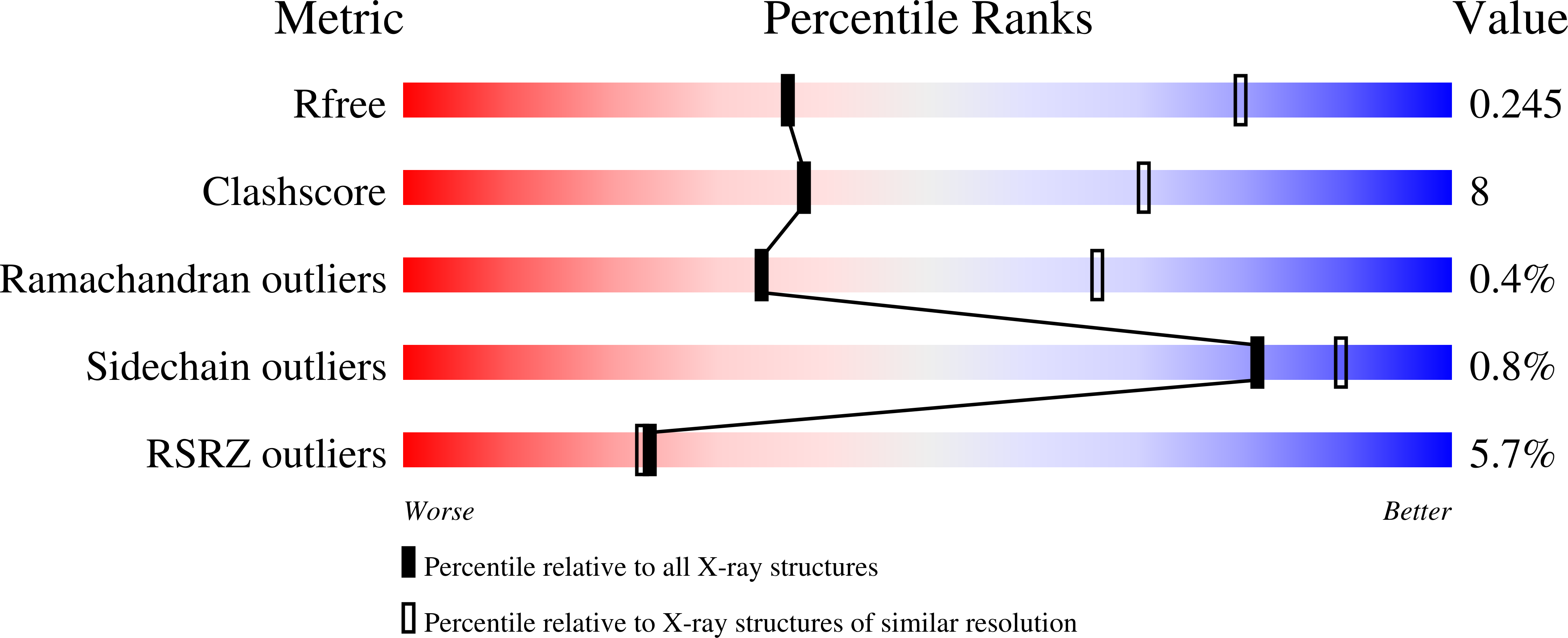
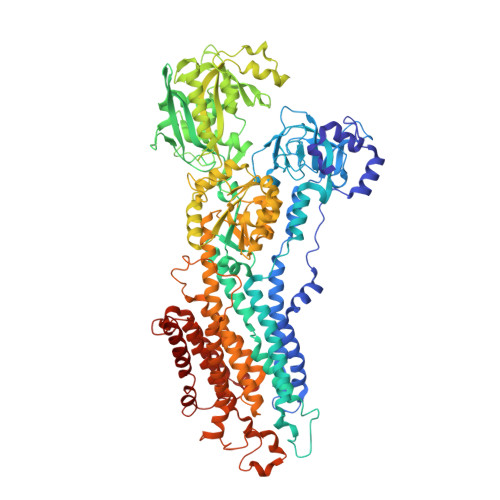
Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase.
Clausen, J.D., Bublitz, M., Arnou, B., Olesen, C., Andersen, J.P., Moller, J.V., Nissen, P.(2016) Structure 24: 617
- PubMed: 27050689
- DOI: https://doi.org/10.1016/j.str.2016.02.018
- Primary Citation of Related Structures:
5A3Q, 5A3R, 5A3S - PubMed Abstract:
Vanadate is the hallmark inhibitor of the P-type ATPase family; however, structural details of its inhibitory mechanism have remained unresolved. We have determined the crystal structure of sarcoplasmic reticulum Ca(2+)-ATPase with bound vanadate in the absence of Ca(2+). Vanadate is bound at the catalytic site as a planar VO3(-) in complex with water and Mg(2+) in a dephosphorylation transition-state-like conformation. Validating bound VO3(-) by anomalous difference Fourier maps using long-wavelength data we also identify a hitherto undescribed Cl(-) site near the dephosphorylation site. Crystallization was facilitated by trinitrophenyl (TNP)-derivatized nucleotides that bind with the TNP moiety occupying the binding pocket that normally accommodates the adenine of ATP, rationalizing their remarkably high affinity for E2P-like conformations of the Ca(2+)-ATPase. A comparison of the configurations of bound nucleotide analogs in the E2¡¤VO3(-) structure with that in E2¡¤BeF3(-) (E2P ground state analog) reveals multiple binding modes to the Ca(2+)-ATPase.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Aarhus University, 8000 Aarhus, Denmark; Centre for Membrane Pumps in Cells and Disease - PUMPKIN, Danish National Research Foundation, Aarhus University, 8000 Aarhus, Denmark; Department of Biomedicine, Aarhus University, 8000 Aarhus, Denmark.