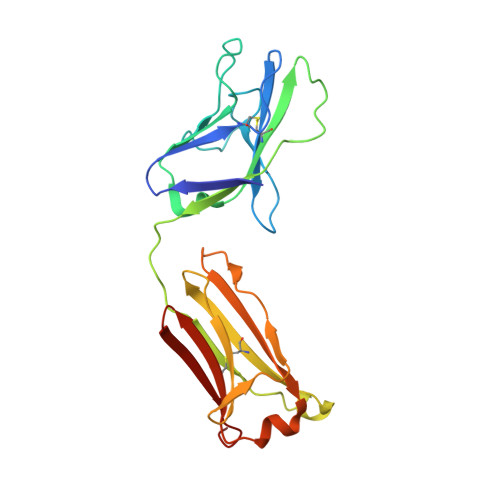
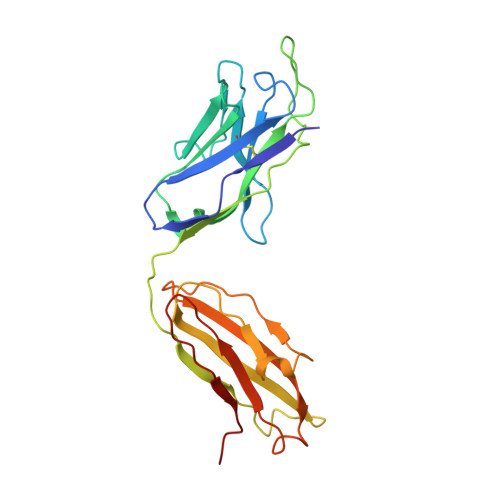
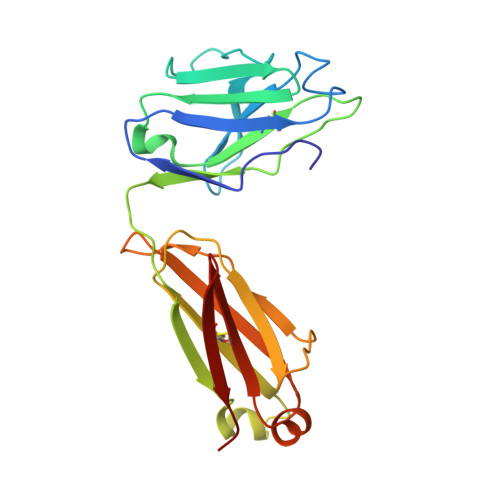
Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site.
Gristick, H.B., von Boehmer, L., West, A.P., Schamber, M., Gazumyan, A., Golijanin, J., Seaman, M.S., Fatkenheuer, G., Klein, F., Nussenzweig, M.C., Bjorkman, P.J.(2016) Nat Struct Mol Biol 23: 906-915
- PubMed: 27617431
- DOI: https://doi.org/10.1038/nsmb.3291
- Primary Citation of Related Structures:
5T3X, 5T3Z - PubMed Abstract:
HIV-1 vaccine design is informed by structural studies elucidating mechanisms by which broadly neutralizing antibodies (bNAbs) recognize and/or accommodate N-glycans on the trimeric envelope glycoprotein (Env). Variability in high-mannose and complex-type Env glycoforms leads to heterogeneity that usually precludes visualization of the native glycan shield. We present 3.5-?- and 3.9-?-resolution crystal structures of the HIV-1 Env trimer with fully processed and native glycosylation, revealing a glycan shield of high-mannose and complex-type N-glycans, which we used to define complete epitopes of two bNAbs. Env trimer was complexed with 10-1074 (against the V3-loop) and IOMA, a new CD4-binding site (CD4bs) antibody. Although IOMA derives from VH1-2*02, the germline gene of CD4bs-targeting VRC01-class bNAbs, its light chain lacks the short CDRL3 that defines VRC01-class bNAbs. Thus IOMA resembles 8ANC131-class/VH1-46-derived CD4bs bNAbs, which have normal-length CDRL3s. The existence of bNAbs that combine features of VRC01-class and 8ANC131-class antibodies has implications for immunization strategies targeting VRC01-like bNAbs.
Organizational Affiliation:
Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, California, USA.