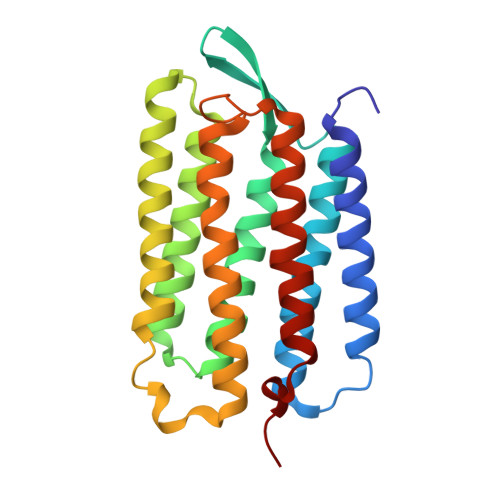
X-ray structure analysis of bacteriorhodopsin at 1.3 angstrom resolution.
Hasegawa, N., Jonotsuka, H., Miki, K., Takeda, K.(2018) Sci Rep 8: 13123-13123
- PubMed: 30177765
- DOI: https://doi.org/10.1038/s41598-018-31370-0
- Primary Citation of Related Structures:
5ZIL, 5ZIM, 5ZIN - PubMed Abstract:
Bacteriorhodopsin (bR) of Halobacterium salinarum is a membrane protein that acts as a light-driven proton pump. bR and its homologues have recently been utilized in optogenetics and other applications. Although the structures of those have been reported so far, the resolutions are not sufficient for elucidation of the intrinsic structural features critical to the color tuning and ion pumping properties. Here we report the accurate crystallographic analysis of bR in the ground state. The influence of X-rays was suppressed by collecting the data under a low irradiation dose at 15?K. Consequently, individual atoms could be separately observed in the electron density map at better than 1.3?? resolution. Residues from Thr5 to Ala233 were continuously constructed in the model. The twist of the retinal polyene was determined to be different from those in the previous models. Two conformations were observed for the proton release region. We discuss the meaning of these fine structural features.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto, 606-8502, Japan.