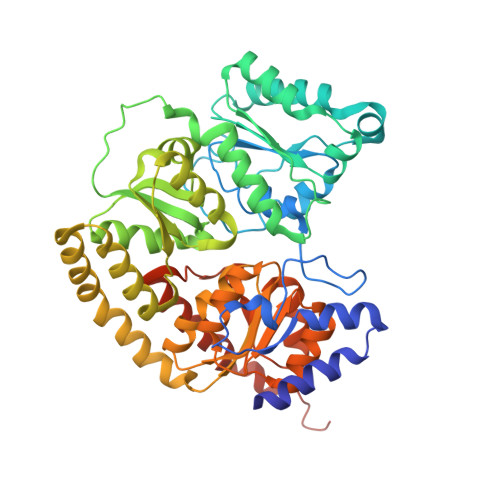
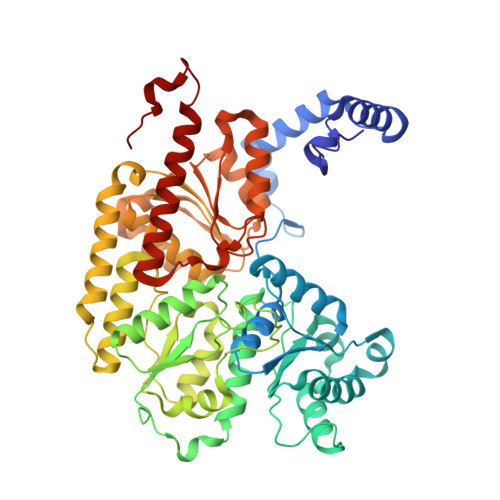
Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Kang, W., Lee, C.C., Jasniewski, A.J., Ribbe, M.W., Hu, Y.(2020) Science 368: 1381-1385
- PubMed: 32554596
- DOI: https://doi.org/10.1126/science.aaz6748
- Primary Citation of Related Structures:
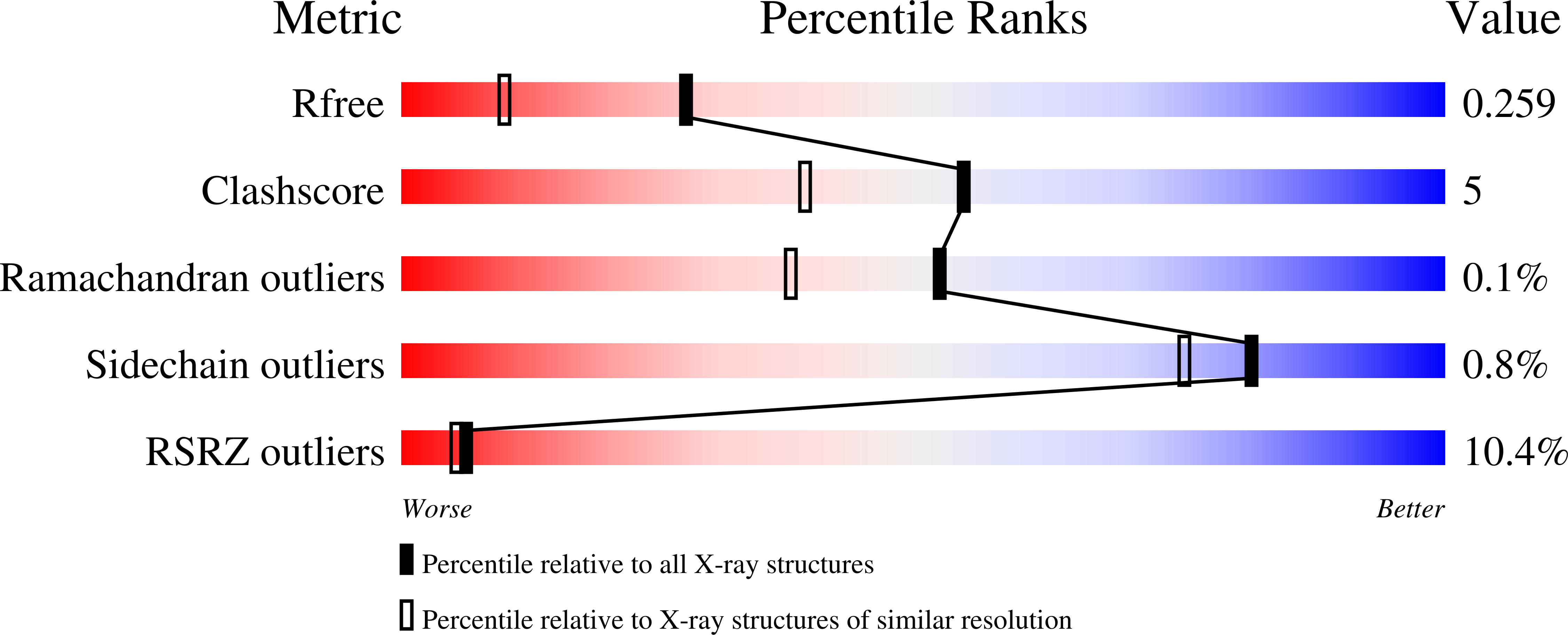
6UG0, 6VXT - PubMed Abstract:
The enzyme nitrogenase uses a suite of complex metallocofactors to reduce dinitrogen (N 2 ) to ammonia. Mechanistic details of this reaction remain sparse. We report a 1.83-angstrom crystal structure of the nitrogenase molybdenum-iron (MoFe) protein captured under physiological N 2 turnover conditions. This structure reveals asymmetric displacements of the cofactor belt sulfurs (S2B or S3A and S5A) with distinct dinitrogen species in the two ¦Á¦Â dimers of the protein. The sulfur-displaced sites are distinct in the ability of protein ligands to donate protons to the bound dinitrogen species, as well as the elongation of either the Mo-O5 (carboxyl) or Mo-O7 (hydroxyl) distance that switches the Mo-homocitrate ligation from bidentate to monodentate. These results highlight the dynamic nature of the cofactor during catalysis and provide evidence for participation of all belt-sulfur sites in this process.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, CA 92697-3900, USA.