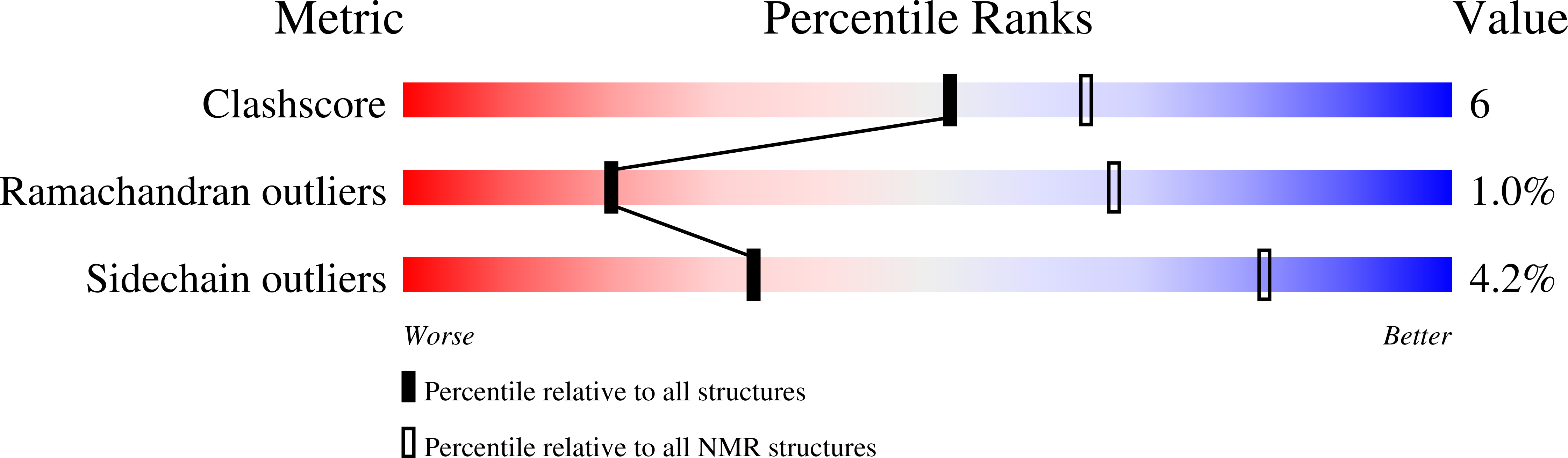Expanding the space of protein geometries by computational design of de novo fold families.
Pan, X., Thompson, M.C., Zhang, Y., Liu, L., Fraser, J.S., Kelly, M.J.S., Kortemme, T.(2020) Science 369: 1132-1136
- PubMed: 32855341
- DOI: https://doi.org/10.1126/science.abc0881
- Primary Citation of Related Structures:
6VG7, 6VGA, 6VGB, 6W90 - PubMed Abstract:
Naturally occurring proteins vary the precise geometries of structural elements to create distinct shapes optimal for function. We present a computational design method, loop-helix-loop unit combinatorial sampling (LUCS), that mimics nature's ability to create families of proteins with the same overall fold but precisely tunable geometries. Through near-exhaustive sampling of loop-helix-loop elements, LUCS generates highly diverse geometries encompassing those found in nature but also surpassing known structure space. Biophysical characterization showed that 17 (38%) of 45 tested LUCS designs encompassing two different structural topologies were well folded, including 16 with designed non-native geometries. Four experimentally solved structures closely matched the designs. LUCS greatly expands the designable structure space and offers a new paradigm for designing proteins with tunable geometries that may be customizable for novel functions.
Organizational Affiliation:
Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, CA, USA. xingjiepan@gmail.com tanjakortemme@gmail.com.