Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Cussol, L., Mauran-Ambrosino, L., Buratto, J., Belorusova, A.Y., Neuville, M., Osz, J., Fribourg, S., Fremaux, J., Dolain, C., Goudreau, S.R., Rochel, N., Guichard, G.(2021) Angew Chem Int Ed Engl 60: 2296-2303
- PubMed: 32935897
- DOI: https://doi.org/10.1002/anie.202008992
- Primary Citation of Related Structures:
6HFA, 6XZH, 6XZI, 6XZJ, 6XZK, 6XZV - PubMed Abstract:
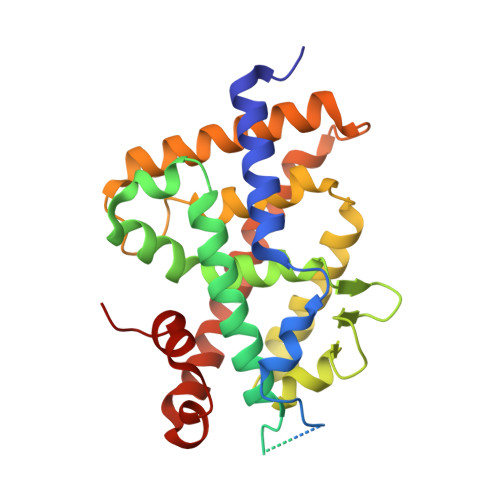
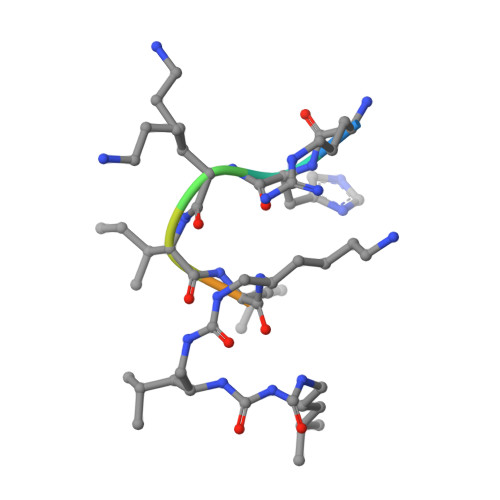
Efficient optimization of a peptide lead into a drug candidate frequently needs further transformation to augment properties such as bioavailability. Among the different options, foldamers, which are sequence-based oligomers with precise folded conformation, have emerged as a promising technology. We introduce oligourea foldamers to reduce the peptide character of inhibitors of protein-protein interactions (PPI). However, the precise design of such mimics is currently limited by the lack of structural information on how these foldamers adapt to protein surfaces. We report a collection of X-ray structures of peptide-oligourea hybrids in complex with ubiquitin ligase MDM2 and vitamin?D receptor and show how such hybrid oligomers can be designed to bind with high affinity to protein targets. This work should enable the generation of more effective foldamer-based disruptors of PPIs in the context of peptide lead optimization.
Organizational Affiliation:
Univ. Bordeaux, CNRS, Bordeaux INP, CBMN, UMR 5248, Institut Europ¨¦en de Chimie et Biologie, 2 rue Robert Escarpit, F-33607, Pessac, France.