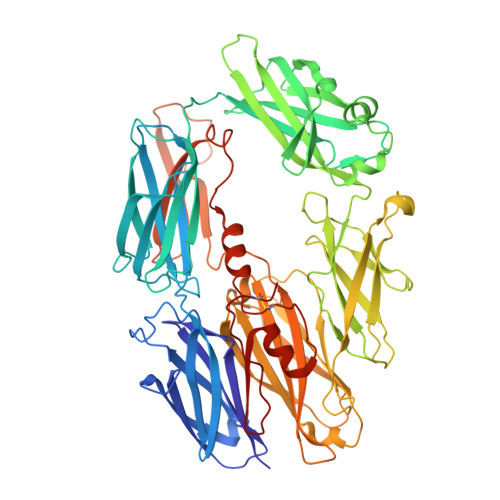
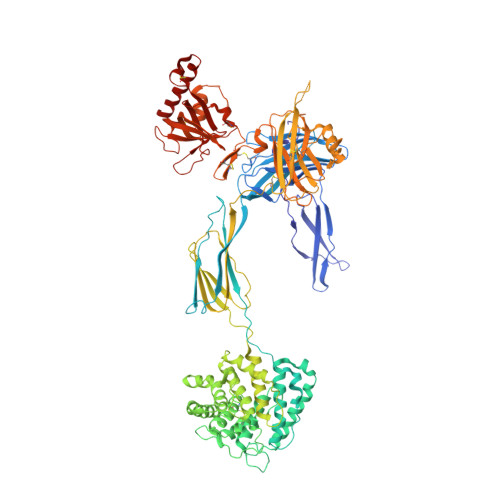
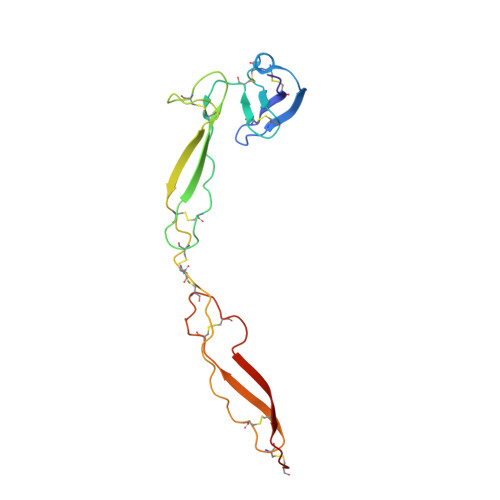
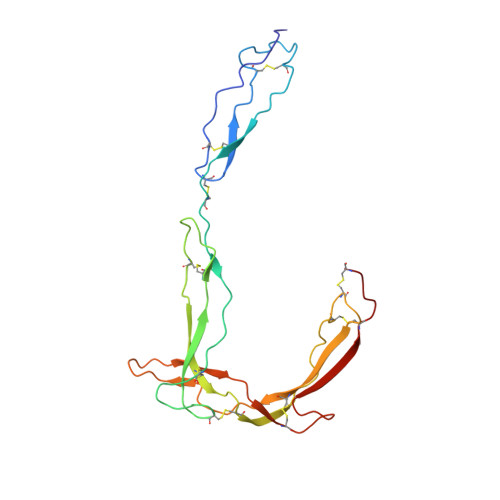
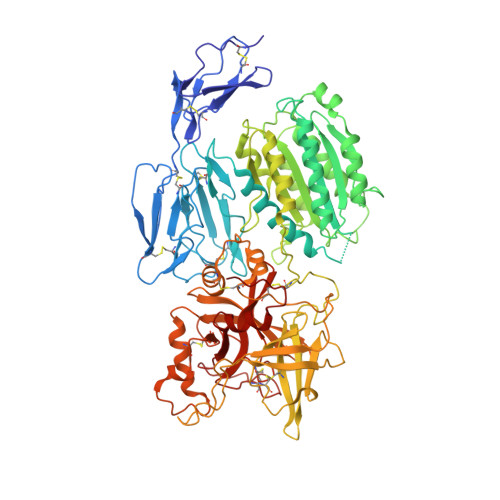
Structure determination of an unstable macromolecular complex enabled by nanobody-peptide bridging.
Lorentzen, J., Pedersen, D.V., Gadeberg, T.A.F., Andersen, G.R.(2022) Protein Sci 31: e4432-e4432
- PubMed: 36173177
- DOI: https://doi.org/10.1002/pro.4432
- Primary Citation of Related Structures:
7NOZ - PubMed Abstract:
Structure determination of macromolecular complexes is challenging if subunits can dissociate during crystallization or preparation of electron microscopy grids. We present an approach where a labile complex is stabilized by linking subunits though introduction of a peptide tag in one subunit that is recognized by a nanobody tethered to a second subunit. This allowed crystal structure determination at 3.9?? resolution of the highly non-globular 320?kDa proconvertase formed by complement components C3b, factor B, and properdin. Whereas the binding mode of properdin to C3b is preserved, an internal rearrangement occurs in the zymogen factor B von Willebrand domain type A domain compared to the proconvertase not bound to properdin. The structure emphasizes the role of two noncanonical loops in thrombospondin repeats 5 and 6 of properdin in augmenting the activity of the C3 convertase. We suggest that linking of subunits through peptide specific tethered nanobodies represents a simple alternative to approaches like affinity maturation and chemical cross-linking for the stabilization of large macromolecular complexes. Besides applications for structural biology, nanobody bridging may become a new tool for biochemical analysis of unstable macromolecular complexes and in vitro selection of highly specific binders for such complexes.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Section for Protein Science, Aarhus Universitet, Aarhus, Denmark.