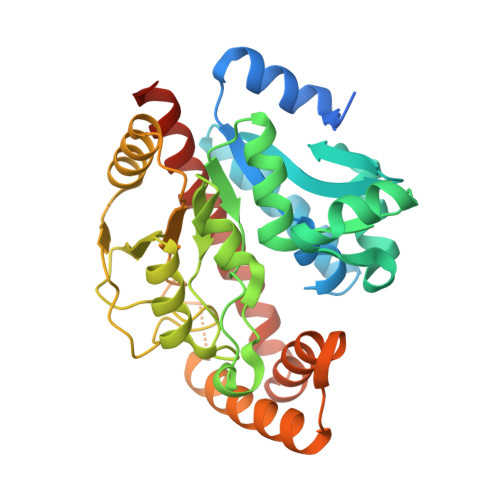
Structure and function of the pseudouridine 5'-monophosphate glycosylase PUMY from Arabidopsis thaliana.
Lee, J., Kim, S.H., Rhee, S.(2024) RNA Biol 21: 1-10
- PubMed: 38117089
- DOI: https://doi.org/10.1080/15476286.2023.2293340
- Primary Citation of Related Structures:
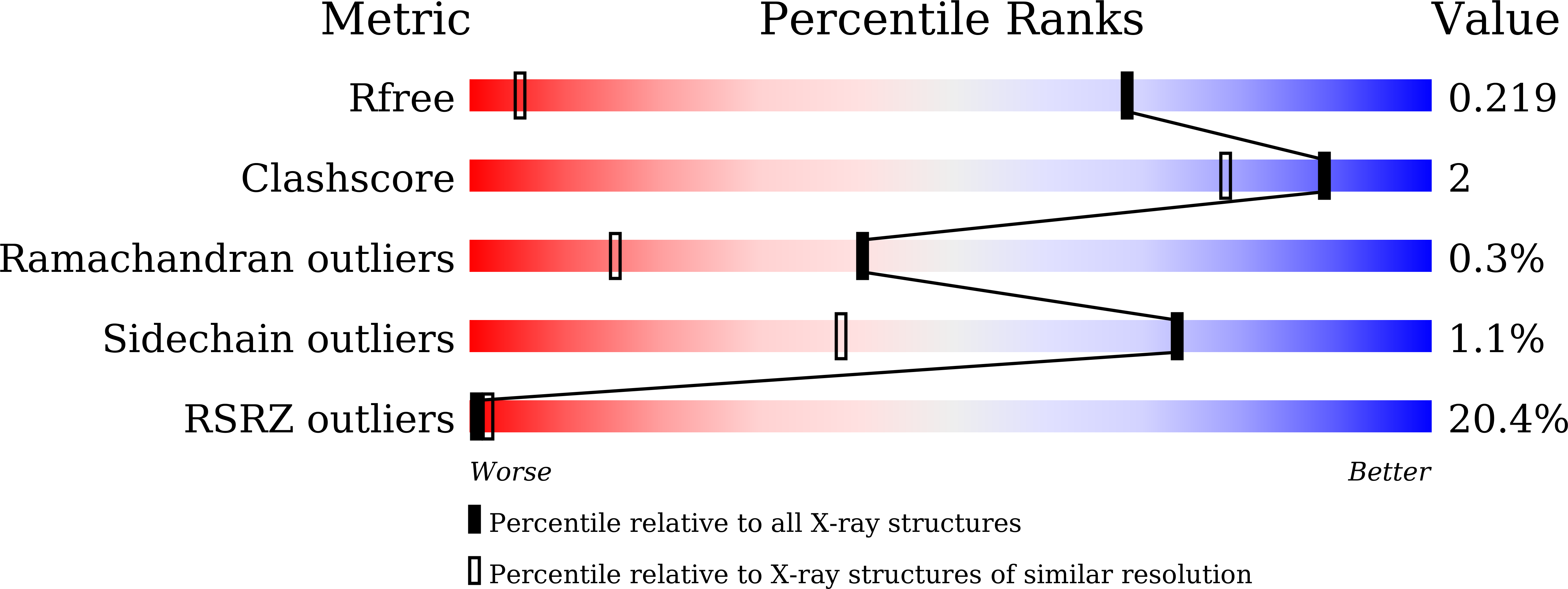
8K05, 8K06, 8K07 - PubMed Abstract:
Pseudouridine is a noncanonical C -nucleoside containing a C-C glycosidic linkage between uracil and ribose. In the two-step degradation of pseudouridine, pseudouridine 5'-monophosphate glycosylase (PUMY) is responsible for the second step and catalyses the cleavage of the C-C glycosidic bond in pseudouridine 5'-monophosphate (¦·MP) into uridine and ribose 5'-phosphate, which are recycled via other metabolic pathways. Structural features of Escherichia coli PUMY have been reported, but the details of the substrate specificity of ¦·MP were unknown. Here, we present three crystal structures of Arabidopsis thaliana PUMY in different ligation states and a kinetic analysis of ¦·MP degradation. The results indicate that Thr149 and Asn308, which are conserved in the PUMY family, are structural determinants for recognizing the nucleobase of ¦·MP. The distinct binding modes of ¦·MP and ribose 5'-phosphate also suggest that the nucleobase, rather than the phosphate group, of ¦·MP dictates the substrate-binding mode. An open-to-close transition of the active site is essential for catalysis, which is mediated by two ¦Á-helices, ¦Á11 and ¦Á12, near the active site. Mutational analysis validates the proposed roles of the active site residues in catalysis. Our structural and functional analyses provide further insight into the enzymatic features of PUMY towards ¦·MP.
Organizational Affiliation:
Department of Agricultural Biotechnology, Seoul National University, Seoul, Korea.