Crystal structure of SARS-CoV-2 nsp16/nsp10 in complex with Cap-1 RNA
Misra, A., Rahisuddin, R., Gupta, Y.K.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 2'-O-methyltransferase | 298 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b EC: 2.1.1 | 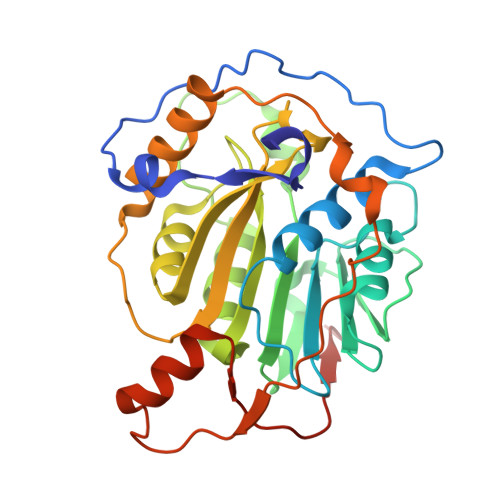 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Non-structural protein 10 | 139 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b | 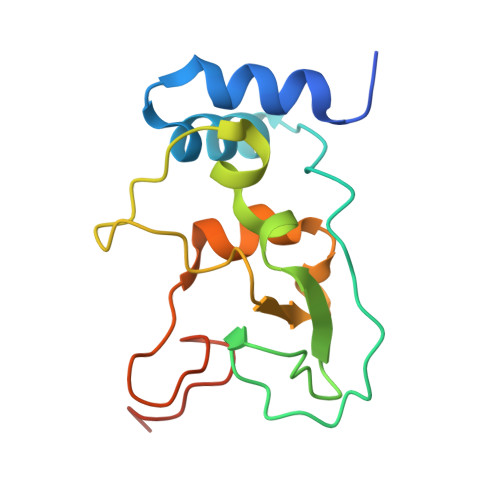 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (5'-R(*AP*UP*UP*AP*AP*AP*GP*GP*UP*U)-3') | 10 | Severe acute respiratory syndrome coronavirus 2 | 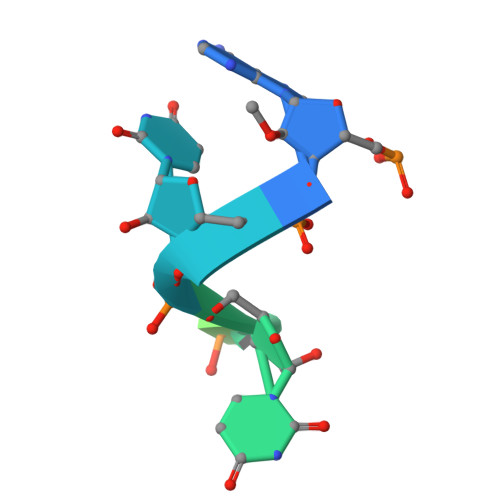 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| M7G Query on M7G | EA [auth E], FA [auth F] | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE C11 H18 N5 O11 P2 SBASPRRECYVBRF-KQYNXXCUSA-O |  | ||
| SAH (Subject of Investigation/LOI) Query on SAH | H [auth A], U [auth C] | S-ADENOSYL-L-HOMOCYSTEINE C14 H20 N6 O5 S ZJUKTBDSGOFHSH-WFMPWKQPSA-N |  | ||
| ZN Query on ZN | BA [auth D], CA [auth D], N [auth B], O [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | AA [auth C] DA [auth D] I [auth A] J [auth A] K [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| MG Query on MG | G [auth A], T [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( ? ) | Angle ( ? ) |
|---|---|
| a = 111.747 | ¦Á = 90 |
| b = 58.893 | ¦Â = 95.481 |
| c = 117.021 | ¦Ă = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | R01AI161363 |