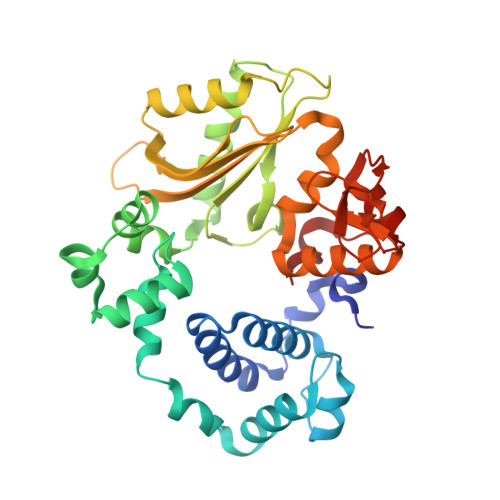
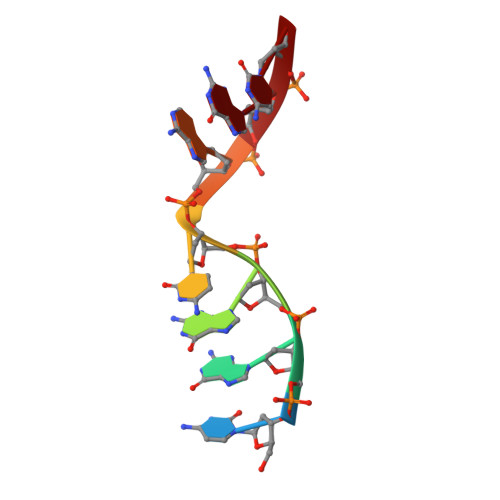
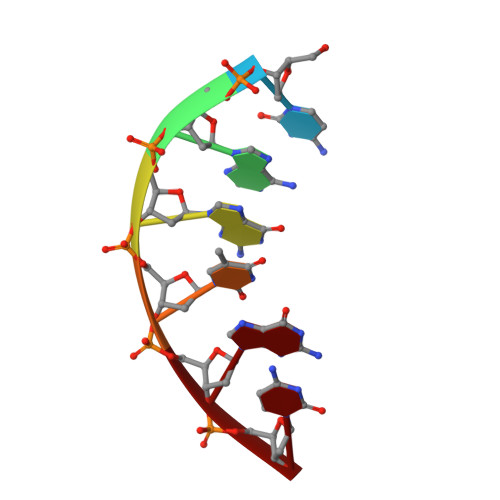
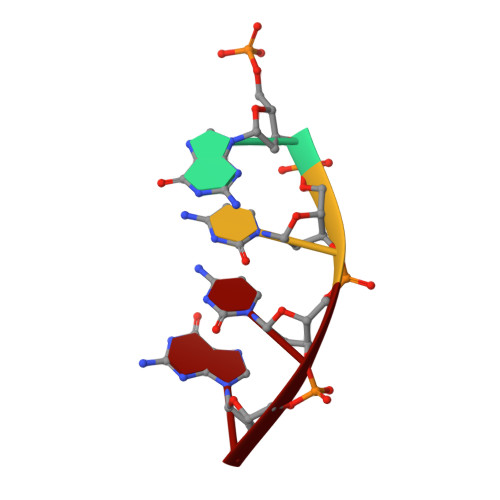
Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Kaminski, A.M., Chiruvella, K.K., Ramsden, D.A., Bebenek, K., Kunkel, T.A., Pedersen, L.C.(2022) Nat Commun 13: 3806-3806
- PubMed: 35778389
- DOI: https://doi.org/10.1038/s41467-022-31278-4
- Primary Citation of Related Structures:
7M07, 7M09, 7M0A, 7M0B, 7M0D, 7M0E - PubMed Abstract:
DNA double-strand breaks (DSBs) threaten genomic stability, since their persistence can lead to loss of critical genetic information, chromosomal translocations or rearrangements, and cell death. DSBs can be repaired through the nonhomologous end-joining pathway (NHEJ), which processes and ligates DNA ends efficiently to prevent or minimize sequence loss. Polymerase ¦Ë (Pol¦Ë), one of the Family X polymerases, fills sequence gaps of DSB substrates with a strict specificity for a base-paired primer terminus. There is little information regarding Pol¦Ë's approach to engaging such substrates. We used in vitro polymerization and cell-based NHEJ assays to explore the contributions of conserved loop regions toward DSB substrate specificity and utilization. In addition, we present multiple crystal structures of Pol¦Ë in synapsis with varying biologically relevant DSB end configurations, revealing how key structural features and hydrogen bonding networks work in concert to stabilize these tenuous, potentially cytotoxic DNA lesions during NHEJ.
Organizational Affiliation:
Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, 111 TW Alexander Dr., Bldg. 101, Research Triangle Park, NC, 27709, USA.