RCSB PDB Help
Exploring a 3D Structure
Genome
Introduction
What is a Genome?
A genome is the complete set of genes and/or genetic material of a cell or organism. It is the blueprint that cells use to make proteins and RNA. In some organisms the entire genome is present as one large DNA molecule while in other organisms it is organized into several chromosomes. Often the genes of proteins that participate in a biological process or pathway are located close to each other so that they can be expressed and/or regulated together.
Why explore Genomes?
Knowing where the gene for the protein/RNA of interest is located in the genome can provide insights into its expression, functions, and regulation. Also, this knowledge can help you understand the impact of any mutations, deletions, or insertions into the introns and intergenic DNA regions on the functions of the protein/RNA of interest.
How is Genome information mapped?
For protein entities in 3D structures [experimental structures or computed structure models (CSMs)] the Protein Information Resource (PIR) provides a mapping between its UniProt identifier (protein sequence) and RefSeq records (collection of genomic DNA, transcript, protein sequences, and various annotations). If this mapping is available, relevant information is included in the Genome tab for that polymer entity. Learn more about Genome view.
Documentation
The Interface
This tab presents information about the gene(s) that code for the protein(s) in the structure.
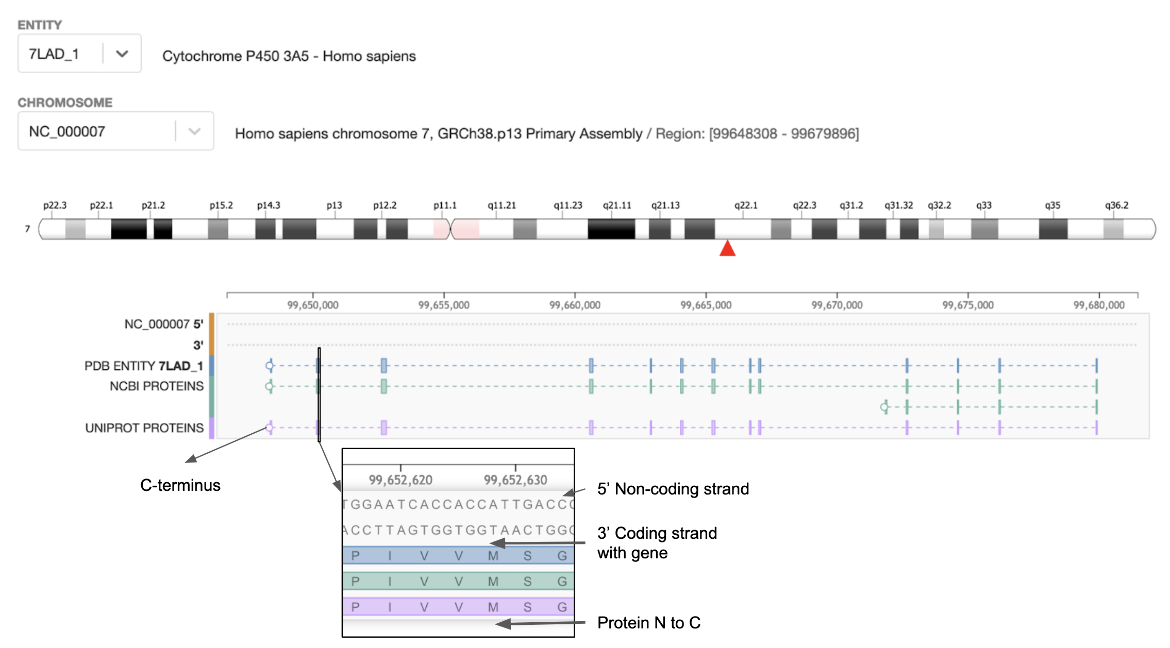
|
| Figure 1: Genome View of the Cytochrome P450 protein for the PDB entry 7lad. The inset shows a zoomed in view of part of an exon and amino acids sequence (from two protein sequence data resources). |
- The top part of the page displays the chromosome and the red arrow marks the location of the gene.
- The gene¡¯s 5¡¯ and 3¡¯ sequences are displayed along with the amino acid sequences of the protein coded by the gene.
Learn more about the conventions used in this display and how to use information presented in this view.
Learning about the Structure
Some of the things that can be learned from the genome view include:
- Translations of gene and protein sequences for the gene of interest
- Presence and location of introns in the gene(s) of interest
- Opportunity to map mutations, insertions, or deletions to the gene¡¯s introns and exons based on information from the literature and/or bioinformatics data resources that have single nucleotide polymorphisms (SNPs) data and other genetic information (e.g., OMIM)
Exploring other Structures
- Knowledge about the location of the gene of interest on the chromosome may help you look up other genes that are located in its neighborhood and understand how their expression and functions might impact the gene or protein of interest.














