RCSB PDB Help
Search and Browse > Advanced Search
Sequence Motif Search
Introduction
What is a sequence motif?
Sequence motifs are short segments of conserved protein or nucleic acid sequences, that are present in many proteins or genes (respectively) and believed to have specific functional significance.
In some cases, the entire set of amino acids or nucleic acids in the sequence is conserved and required to perform the specific function.
In other cases, only amino acids or nucleic acids at specific locations in the sequence motif may be conserved and significant for the function.
What is a Sequence Motif Search?
The sequence motif search option allows you to query for amino acid or nucleotide sequence fragments in an FASTA sequence that appear frequently in polymers present in 3D structures.
Why run a Sequence Motif Search?
Finding a specific sequence motif in a protein or nucleic acid suggests that it may have the function associated with the motif; i.e., it can be used to predict function(s).
Another reason to run sequence motif searches is that it is indeed different from a regular similarity-based sequence search (e.g., BLAST) in two ways:
- The sequence defining the sequence motif is short (so the similarity-based searches will not work effectively)
- Parts of the sequence motif may have alternate sequences or may not be conserved at all (so specific conditions have to be included in the query for defining the non-contiguous conserved amino acids/nucleotides in the sequence motif)
Running a search
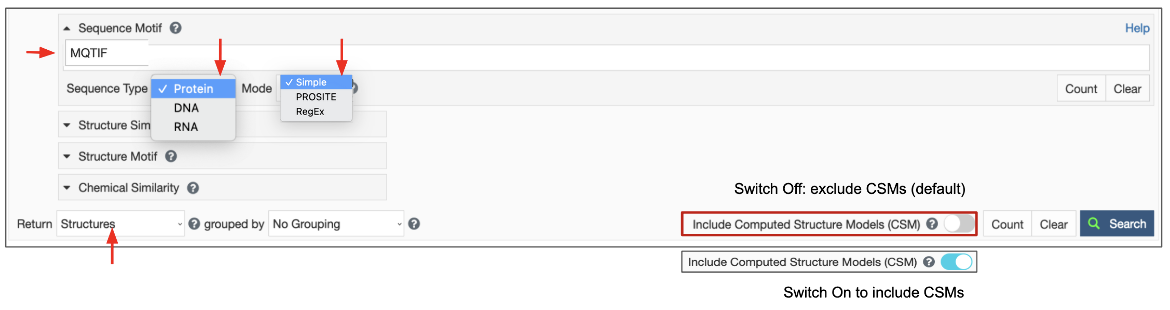
The sequence motif search options are available from the Advanced Search Query builder (Figure 1).
Query options
-
Set the Sequence Type to search for protein (amino acid), DNA, or RNA sequences.
-
Set the Mode to specify the motif syntax as simple, PROSITE, or regex.
-
Set the Data Source to search among experimental records only, computational records, or both.
Sequence Type
In all three Modes, amino acid residue (or nucleotide) types are specified using one-letter codes, which are defined by
IUPAC.
For example: for amino acid sequences, R is arginine; for RNA sequences, U is uracil.
Nucleotide sequences also support so-called ambiguous codes; for example, S is either cytosine or guanine.
Only Simple and PROSITE Modes support ambiguous codes.
Below is a full reference of one-letter codes.
Queries are case-insensitive for all three Modes:
ATGC and atgc are identical.
(This also applies to X and x in Simple and PROSITE Modes.)
? Tables of one-letter codes
| code | meaning |
|---|---|
A | adenine |
C | cytosine |
G | guanine |
T 1 | thymine |
U 1 | uracil |
B 2 | C/G/T/U |
D 2 | A/G/T/U |
H 2 | A/C/T/U |
K 2 | G/T | /
M 2 | A/C |
R 2 | A/G |
S 2 | C/G |
V 2 | A/C/G |
W 2 | A/T/U |
Y 2 | C/T/U |
N 2 | any base |
1
T is restricted to DNA;
U is restricted to RNA
3 Termed ambiguous; only supported in Simple and PROSITE Modes.
| code | meaning |
|---|---|
A | alanine |
C | cysteine |
D | aspartic acid |
E | glutamic acid |
F | phenylalanine |
G | glycine |
H | histidine |
I | isoleucine |
K | lysine |
L | leucine |
M | methionine |
N | asparagine |
P | proline |
Q | glutamine |
R | arginine |
S | serine |
T | threonine |
V | valine |
W | tryptophan |
Y | tyrosine |
Simple Mode
Input a sequence of one or more of one-letter codes.
Ambiguous nucleotide codes are supported,
and the wildcard symbol (X) can be used to represent any amino acid or nucleotide.
Use < and > to match the N- and C-termini, respectively.
Examples
-
XPPXP(protein): SH3 domains (any ¡ú proline ¡ú proline ¡ú any ¡ú proline) -
YYY(DNA): 3¡Á cytosine/thymine -
<SSS: any sequence that starts with 3¡Á serine
PROSITE Mode
Complex queries can be expressed using PROSITE patterns.
A PROSITE pattern is composed of one or more atoms,
optionally separated by hyphens (-).
The sequence is optionally terminated by a period (.).
X can be used to stand in for any amino acid or nucleotide type,
and ambiguous nucleotide codes (e.g., B) are supported.
Atom types
Each atom is one of seven types:
- Literal
-
A one-letter code (e.g.,
A). This matches exactly 1 residue. - Any-of (
[]) -
One or more codes enclosed in
[], such as[ATC]. This matches exactly 1 residue whose code is listed. - None-of (
{}) -
One or more codes enclosed in
{}, such as{ATC}. This matches exactly 1 residue whose code is not listed. - N-terminus (
<) -
An N-terminal marker,
<, indicating the start of the sequence. If included, this must be the first element. - C-terminus (
>) -
A C-terminal marker,
>, indicating the end of the sequence. If included, this must be the last element. - Any-of / C-terminus (e.g.,
[A>]) -
A variable C-terminal element, such as
[>AC],[A>C], or[AC>](equivalently). This matches either the end of the sequence or exactly 1 reside among those listed (but not both).
Quantifiers
Each literal, wildcard, any-of, and none-of element may be followed by a quantifier
to match the preceding element some number of times.
The quantifier is enclosed in () and can be
Exact, Minimum, or Range:
- Exact
A(2)matches exactly AA.- Minimum
A(2,)matches at AA, AAA, ¡ .- Range
A(2,4)matches AA, AAA, and AAAA.
Regex Mode
Regular expressions (regex) are also supported. This option is more powerful than PROSITE and may be familiar to programmers. Note that the service may refuse to process some queries.
A regex pattern contains one or more atoms, each with an optional quantifier.
| denotes a logical or, and () groups atoms into groups.
Ambiguous nucleotide codes are not supported, nor is X.
Use . instead of X,
and use [CGT] (for DNA) or [CGU] (for RNA)
instead of B.
Examples
-
W.{7}G.{20}Lmatches tryptophan ¡ú 7¡Áany ¡ú glycine ¡ú 20¡Áany ¡ú lysine. -
C.{2,4}C.{12}H.{3,5}Hmatches the zinc finger motif that binds Zn in a DNA-binding domain. -
^H+$matches N-terminus ¡ú 1+ histidine ¡ú C-terminus. -
[AG].{4}GK[ST]matches the Walker (P loop) motif that binds ATP or GTP.
Viewing results
Pre-search checklist
- Before running the search remember to do the following:
- change the result return option to Polymer entities
- decide whether to include CSMs (default) or exclude them (by turning off the toggle switch next to the Search button.
Result options
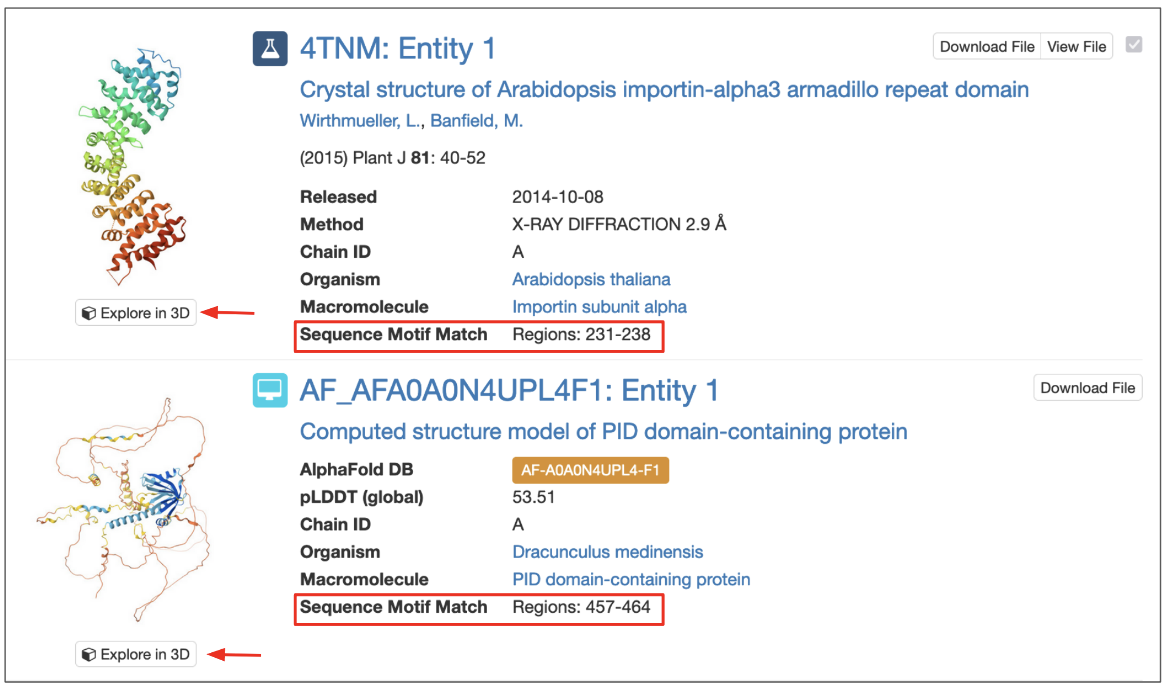
The search results display the numbering for the sequence match region (corresponding to PDBx/mmCIF file numbering) (Figure 2).
Click on the 3D View button included for each matched result to view the structure interactively in 3D.
The matched region specified in the results can be examined closely.
Example searches
- Query for SH3 domains ¨C use the Simple Mode query
XPPXP, whereXis any residue andPis proline. - Query for a specific pattern of sequence ¨C use the PROSITE Mode query
[AC]-x-V-x(4)-{ED}, which translates to [Ala or Cys]-any-Val-any-any-any-any-{any but Glu or Asp}. - Query for the Walker (P loop) motif that binds ATP or GTP ¨C use the Regex Mode query
[AG]....GK[ST], where A or G is followed by 4 variable residues, then G and K, and finally S or T.
Extended (advanced) information
PROSITE Mode details
Terminology
This documentation uses the following definitions.
- Atom
-
A PROSITE syntax item to match against 1 residue or nucleotide.
A literal (e.g.,
A), gap (.), any-of (e.g.,[CG]), none-of (e.g.,{AT}, N-terminus (<), C-terminus (>[>AT]) - Term
- An atom with its quantifier, if any
Nonstandard but allowed in RCSB
RCSB PROSITE is more forgiving than standard PROSITE; it differs in the following ways:-
Case is ignored.
Aandaare the same, as areXandx. -
Range quantifiers (
(x,y)) may be used for all atoms, not just gaps (x). For example,A(1,4)matches between 1 and 4 alanines. In contrast, standard PROSITE only permits, for example,x(1,4). -
Hyphens (
-) may be omitted, even with one-letter nucleotide codes, such asB. Hyphens are ignored as long as they are used in valid positions.
RCSB-specific rules
Some parts of the PROSITE specification can be interpreted in multiple ways. RCSB PROSITE has decided on these rules:-
Spaces (characters with Unicode category Zs) are ignored when in reasonable positions.
For example,
A T{1, 3}is allowed. -
The query must contain at least 1 atom.
(
<,>,<>, and the empty string are forbidden.) -
Any-of matches (
[]) require at least 1 character. -
None-of matches (
{}) cannot include every one-letter code.{ATGC}is invalid for DNA sequences (and could never match a sequence). -
An exact quantifier
(n)is allowed if and only ifn ¡Ý 1. -
A range quantifier
(m, n)is allowed if and only ifn ¡Ý mandm > 0.
Formal grammar
This grammar uses RFC 5234 ABNF.
query = start *(['-'] term) ['-' end] ['.']
; ^ ^ ^
; required 0 or more optional
start = term / (nterm non-gap-term) / (nterm gap)
% EITHER: A term (1+ elements) without an N-term
% OR: N-term with non-gap term (1+ elements)
% OR: single gap (1 element; non-repeated)
end = term / (non-gap-term / cterm) / (gap cterm)
term = element [count / range]
element = code / any-of / none-of / gap
non-gap-term = non-gap-element [count / range]
non-gap-element = code / any-of / none-of
aa = "a one-letter code"
gap = 'x'
; Matches any single residue
any-of = '[' 1*aa ']'
; Matches any single residue included in []
; For example, [ACE] matches A, C, or E
none-of = '{' 1*aa '}'
; Matches any single residue NOT included in {}
count = '(' natural ')'
; An exact number of times to repeat the preceding element
; For example, [AW](3) is equivalent to [AW][AW][AW]
range = '(' number ',' natural ')'
; A min and max number of times to repeat the preceding element
; For example, A{1,3} matches A, AA, and AAA
; Note: min must be less than max
nterm = '<'
; Matches the sequence start (N-terminus)
cterm = cterm-literal / cterm-or-any-of
cterm-literal = '>'=
; Matches the sequence end (C-terminus)
cterm-or-any-of = ('[' (1*aa '>' *aa) / (*aa '>' 1**aa) ']')
; Matches either the sequence end (C-terminus),
; OR an aa included in [] / an aa not included in {}
; For example, [A>] matches either the sequence end or A.
; Valid examples: [A>], [>A], [A,C], [ACDE>]
; Invalid examples: [A>>], [A>C>], [>], []
number = 1*DIGIT
natural = NONZERO *DIGIT
NONZERO = %x31-39
Regex Mode details
Supported and non-supported constructs
The query syntax is IEEE POSIX Extended Regular Expressions. Nearly all of the standard is supported, including advanced constructs like lookarounds and backreferences.
However, a few things are not supported.
Most notably, characters that are not in one-letter codes are not allowed in literals or character classes.
For example, Z and [A-Z] would result in an error.
Named character classes, such as \s and \p{Alpha}, are also not supported.
Queries that will be rejected
In addition, the service will not allow expressions that could seriously degrade performance. Specifically, these are expressions with non-polynomial worst-case runtime or space complexity. The service will reject:- patterns that use non-possessive, inexact quantifiers on groups that match a variable number of characters, n, n > 1;
- patterns that use quantifiers on groups satisfying certain (other) ways;
- patterns that use lazy, inexact quantifiers excessively or in certain ways;
- patterns that use lookarounds excessively or in certain ways;
- patterns with non-polynomial worst-case runtime or memory requirements; and
- patterns with excessive total complexity.
Tips to simplify queries
Follow these guidelines to avoid a query being rejected.
- Do not use lazy quantifiers.
- Avoid lookarounds.
- When applying quantifiers to groups, make sure the group is simple and only use either greedy
?or (preferably) a possessive quantifier. - Use possessive quantifiers where possible.
- Do not begin or end a sequence with
.*,^.*,.*$, or similar.